Compositions and methods for adoptive immunotherapy
阅读说明:本技术 用于过继免疫疗法的组合物和方法 (Compositions and methods for adoptive immunotherapy ) 是由 杨文� 于 2020-07-22 设计创作,主要内容包括:本发明涉及赋予和/或增加由细胞免疫疗法介导的免疫应答的药剂、组合物和方法。(The present invention relates to agents, compositions and methods for conferring and/or increasing immune responses mediated by cellular immunotherapy.)
1. An antigen receptor comprising (I) a first polypeptide chain comprising an extracellular domain comprising a T Cell Receptor (TCR) β chain or an antigen-binding fragment thereof, a transmembrane domain, and a cytoplasmic domain, and (II) a second polypeptide chain comprising an extracellular domain comprising a TCR α chain or an antigen-binding fragment thereof, wherein
Said TCR β chain and said TCR α chain forming an antigen binding site, and
said cytoplasmic domain of said first polypeptide chain or of said second polypeptide chain comprises:
0. 1 or 2 copies of the cytoplasmic domain of human 4-1BB or a fragment thereof, or
0. 1 or 2 copies of the cytoplasmic domain of human CD3 zeta or a fragment thereof, or
0. 1 or 2 copies of the cytoplasmic domain of human CD3 epsilon or a fragment thereof, or
0. 1 or 2 copies of the cytoplasmic domain of human CD28 or a fragment thereof.
2. The antigen receptor of claim 1, wherein the first polypeptide chain is substantially different from the second polypeptide chain in one or more of the transmembrane domain and the cytoplasmic domain.
3. The antigen receptor of claim 1, comprising 1 or 2 copies of the cytoplasmic domain of human 4-1 BB.
4. The antigen receptor of claim 3, comprising 1 or 2 copies of the cytoplasmic domain of human CD3 ζ.
5. The antigen receptor of claim 3 or 4, comprising 1 or 2 copies of the cytoplasmic domain of human CD 28.
6. The antigen receptor of claim 1 or 2, comprising only 1 copy of the cytoplasmic domain of human 4-1BB, 1 copy of the cytoplasmic domain of human CD28, and 1 copy of the cytoplasmic domain of human CD3 ζ.
7. The antigen receptor of claim 1 or 2, comprising only 1 copy of the transmembrane domain of CD28, 1 copy of the transmembrane domain of CD8, 1 copy of the cytoplasmic domain of human 4-1BB, 1 copy of the cytoplasmic domain of human CD28, and 1 copy of the cytoplasmic domain of human CD3 ζ.
8. The antigen receptor of claim 1 or 2, comprising 2 copies of the cytoplasmic domain of human 4-1BB, 2 copies of the cytoplasmic domain of human CD28, and 2 copies of the cytoplasmic domain of human CD3 ζ.
9. The antigen receptor of claim 8, wherein
Said cytoplasmic domain of said first polypeptide chain comprises 1 copy of said cytoplasmic domain of human 4-1BB, 1 copy of said cytoplasmic domain of human CD28, and 1 copy of said cytoplasmic domain of human CD3 ζ, and
the cytoplasmic domain of the second polypeptide chain comprises 1 copy of the cytoplasmic domain of human 4-1BB, 1 copy of the cytoplasmic domain of human CD28, and 1 copy of the cytoplasmic domain of human CD3 zeta.
10. The antigen receptor according to claim 1 or 2, wherein
Said first polypeptide chain comprises 1 copy of said transmembrane domain of human CD28, 1 copy of said cytoplasmic domain of human CD28, and 1 copy of said cytoplasmic domain of human CD3 ζ, and
said second polypeptide chain comprises 1 copy of said transmembrane domain of human CD8 and 1 copy of said cytoplasmic domain of human 4-1 BB.
11. The antigen receptor according to claim 1 or 2, wherein
Said first polypeptide chain comprises 1 copy of said transmembrane domain of human CD8, 1 copy of said cytoplasmic domain of human CD28, and 1 copy of said cytoplasmic domain of human CD3 ζ, and
said second polypeptide chain comprises 1 copy of said transmembrane domain of human CD8, 1 copy of said cytoplasmic domain of human 4-1BB, and 1 copy of said cytoplasmic domain of human CD3 ζ.
12. The antigen receptor according to claim 1 or 2, wherein
Said first polypeptide chain comprises 1 copy of said transmembrane domain of human CD8, 2 copies of said cytoplasmic domain of human CD28, and 1 copy of said cytoplasmic domain of human CD3 ζ, and
said second polypeptide chain comprises 1 copy of said transmembrane domain of human CD8, 2 copies of said cytoplasmic domain of human 4-1BB, and 1 copy of said cytoplasmic domain of human CD3 ζ.
13. The antigen receptor of any one of claims 1-12, wherein the transmembrane domain of the first or second polypeptide chain comprises one selected from the group consisting of: the transmembrane domain of CD8 and the transmembrane domain of CD 28.
14. The antigen receptor of any one of claims 1-13, wherein the antigen binding site binds to a tumor antigen, or a Tumor Associated Antigen (TAA), or a viral antigen in the context of an mhc (hla) restricted manner, or wherein the extracellular domain, when expressed on a cell, binds to a tumor antigen, or a TAA, or a viral antigen in the context of an mhc (hla) restricted manner.
15. The antigen receptor of claim 14, wherein the tumor antigen is NY-ESO-1.
16. A fusion protein comprising a first polypeptide chain and a second polypeptide chain of any one of claims 1-15, wherein the first polypeptide chain and the second polypeptide chain are connected by a protein linker sequence or a self-cleaving peptide sequence.
17. The fusion protein of claim 16, wherein the self-cleaving peptide sequence comprises a P2A, E2A, F2A, or T2A sequence.
18. An isolated nucleic acid or a set of isolated nucleic acids encoding the antigen receptor of any one of claims 1-15 or the fusion protein of claims 16-17.
19. The isolated nucleic acid or a group of isolated nucleic acids of claim 18, comprising
(I) A first nucleic acid sequence encoding said first polypeptide chain comprising a first polypeptide segment of at least 10 amino acids in length, and
(II) a second nucleic acid sequence encoding said second polypeptide chain comprising a second polypeptide segment of at least 10 amino acids in length, wherein
The first polypeptide segment is at least 90% identical to the second polypeptide segment, and
the first and second nucleic acid sequences comprise at least one different codon within a codon encoding the same amino acid residue in the first and second polypeptide segments.
20. The isolated nucleic acid of claim 19, wherein (a) the first polypeptide chain and the second polypeptide chain comprise the same polypeptide sequence of greater than 10 amino acids in length; and (b) the first nucleic acid sequence and the second nucleic acid sequence comprise at least one different codon within codons of the first nucleic acid sequence and the second nucleic acid sequence encoding the same polypeptide sequence.
21. The isolated nucleic acid of any one of claims 19-20, wherein at least 2% of the codons are different.
22. The isolated nucleic acid of any one of claims 19-21, wherein the first nucleic acid sequence and the second nucleic acid sequence are less than 98% identical within the codon.
23. The isolated nucleic acid of any one of claims 19-22, wherein the first nucleic acid sequence and the second nucleic acid sequence contain at least one different codon within a codon of one or more selected from the group consisting of seq id no:
(a) the TCR beta chain or antigen binding fragment thereof,
(b) the TCR alpha chain or antigen binding fragment thereof,
(c) the transmembrane domain of human CD8,
(d) the transmembrane domain of human CD28,
(e) the cytoplasmic domain of human CD3 ζ or fragment thereof,
(f) the cytoplasmic domain of human CD3 epsilon or a fragment thereof,
(g) the cytoplasmic domain of human CD28 or a fragment thereof, and
(h) the cytoplasmic domain of human 4-BB or a fragment thereof.
24. The isolated nucleic acid of claim 23, wherein the isolated nucleic acid encodes one selected from the group consisting of: SEQ ID NO: 9. 64, 10, 11 and 32-41.
25. The isolated nucleic acid of claim 24, wherein the isolated nucleic acid comprises a sequence selected from the group consisting of: SEQ ID NO: 12. 65, 13, 14 and 54-63.
26. The isolated nucleic acid of any one of claims 19-24, wherein the first nucleic acid sequence and the second nucleic acid sequence are linked by a nucleic acid sequence comprising an Internal Ribosome Entry Site (IRES), IRESDNA fragment.
27. A vector comprising the isolated nucleic acid of any one of claims 18-26.
28. The vector of claim 27, wherein the vector is an expression vector.
29. The vector of claim 28, wherein the vector is a viral vector.
30. A cell comprising the antigen receptor of any one of claims 1-15, or the fusion protein of claims 16-17, or the isolated nucleic acid of any one of claims 18-26, or the vector of any one of claims 27-29.
31. The cell of claim 30, wherein the cell is a T cell.
32. A pharmaceutical composition comprising (i) the isolated nucleic acid of any one of claims 18-26, or the vector of any one of claims 27-29, or the cell of any one of claims 30-31, and (ii) a pharmaceutically acceptable carrier.
33. A method of treating a neoplastic or viral infectious disease comprising administering to a subject in need thereof an effective amount of the pharmaceutical composition of claim 32.
Technical Field
The present invention relates to recombinant antigen receptors and uses thereof. T cells engineered to express such antigen receptors are useful in the treatment of diseases characterized by expression of one or more antigens that bind to the antigen receptor.
Background
Adoptive immunotherapy involves the administration of immune effector cells to a patient to produce a therapeutic effect. The advent of Chimeric Antigen Receptor (CAR) T cells provides a useful tool for improving adoptive immunotherapy. To this end, antigen-targeting receptors defining specificity are inserted into T cells using genetic engineering, which greatly expands the ability of adoptive immunotherapy. CAR is a type of antigen-targeting receptor consisting of an intracellular T cell signaling domain fused to an extracellular antigen-binding domain. Antibody-based CARs recognize cell surface antigens directly, without relying on Major Histocompatibility Complex (MHC) -mediated presentation, thereby allowing the use of a single receptor construct specific for any given antigen in all patients. Initially, the CAR fused the antigen recognition domain to the CD3 ζ (CD3 ζ) activation chain of the T Cell Receptor (TCR) complex. Subsequent CAR iterations included secondary costimulatory signals in tandem with CD3 ζ, including the endodomain from CD28 or various TNF receptor family molecules, such as 4-1BB (CD137) and OX40(CD 134). In addition to CD3 ζ, the third generation receptor includes two distinct costimulatory signals, most commonly from CD28 and 4-1 BB. Second and third generation antibody-based CARs improved antitumor efficacy both in vitro and in vivo.
Transgenic T Cell Receptors (TCR) and TCR-based CARs (TCR-CARs) differ from antibody-based CARs primarily in that the TCR and TCR-based CARs bind to antigen in an mhc (hla) restricted manner. Thus, tTCR and TCR-based CARs greatly expand the list of possible targets. Second generation TCR-CAR comprising cytoplasmic domains of human CD28 and CD3 ζ (CD3Z) or human CD28 and CD3 ε (CD3E) established as two separate viral vectors have been described previously (Govers et al, Journal of Immunology,2014,193: 5315-5326), as well as second generation TCR-CAR comprising cytoplasmic domains of human CD28 and CD3 established as a single viral vector as described previously in U.S. Pat. Nos. 9,206,440 and Im EJ et al, registration-deletion gene homology vectors in retroviruses is Suppressed view a substrate of generation code organization. molecular Therapy-Methods & Clinical Development (2014) arm number: 14022. Furthermore, second generation TCR-CARs comprising a single chain TCR and the cytoplasmic domain of human CD28 and CD3 zeta established as a single viral vector have been described previously (Walseng et al, A TCR-based nucleic acid receptor. Sci Rep.2017Sep 6; 7(1): 10713). In addition to CD3 ζ, the third generation receptor includes two distinct costimulatory signals, most commonly from CD28 and 4-1 BB. Second and third generation antibody-based CARs improved antitumor efficacy in vitro and in vivo.
Unlike second or third generation antibody-based CARs, ttcrs and TCR-based CARs face various obstacles, such as loss of stability for efficient tTCR or TCR-based CAR expression and activity. Despite the lack of costimulatory signaling by itself in its own molecule, the presently reported TCR-based CARs lack sufficient costimulatory signaling elements (e.g., 4-1BB) or lack the optimal design for optimal cell surface expression cassette CAR-mediated T cell activity. There is a need for new TCR-based CAR designs and adoptive therapies that provide cells with enhanced function.
Disclosure of Invention
The present disclosure addresses the above-mentioned needs in a number of respects. In one aspect, the present disclosure provides an antigen receptor comprising (I) a first polypeptide chain comprising an extracellular domain comprising a TCR β chain or an antigen-binding fragment thereof, a transmembrane (TM or TM) domain (TMD), and a cytoplasmic domain (Cyt or Cyt); and (II) a second polypeptide chain comprising an extracellular domain (Ec) comprising a TCR a chain or an antigen-binding fragment thereof, a transmembrane domain, and a cytoplasmic domain. The TCR β chain and the TCR α chain form an antigen binding site. In embodiments, the first polypeptide chain is substantially different from the second polypeptide chain in one or more of the transmembrane domain and the cytoplasmic domain.
The cytoplasmic domain of the first polypeptide chain or the cytoplasmic domain of the second polypeptide chain comprises (a)0, 1, or 2 copies of the cytoplasmic domain of human 4-1BB or fragment thereof, or (b)0, 1, or 2 copies of the cytoplasmic domain of human CD3 ζ (CD3Z) or fragment thereof, or (c)0, 1, or 2 copies of the cytoplasmic domain of human CD3 ∈ (CD3E) or fragment thereof, or (d)0, 1, or 2 copies of the cytoplasmic domain of human CD28 or fragment thereof.
In the antigen receptor, the transmembrane domain of the first polypeptide chain or the transmembrane domain of the second polypeptide chain may comprise one selected from: the transmembrane domain of CD8 and the transmembrane domain of CD 28.
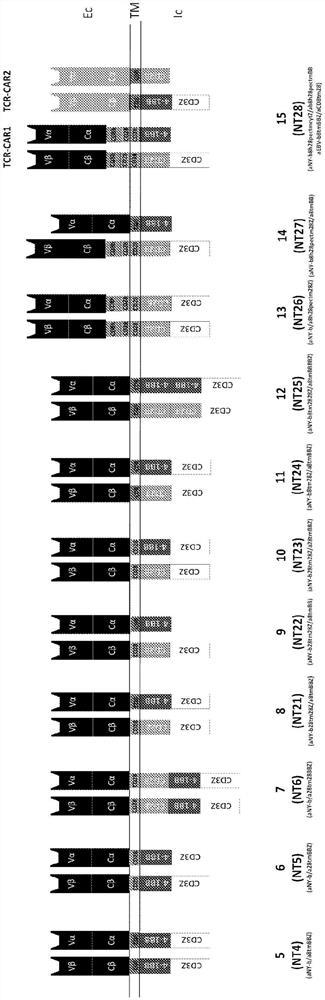
In some embodiments, the antigen receptor comprises 1 or 2 copies of the cytoplasmic domain of human 4-1 BB. Examples of such antigen receptors include those encoded by the vectors NT4, 5,6, 21, 22, 23, 24, 25 and 27 described herein. In some examples, the antigen receptor may comprise 1 or 2 copies of the cytoplasmic domain of human CD3 ζ. In other examples, the antigen receptor may comprise 1 or 2 copies of the cytoplasmic domain of human CD 28. Examples include those encoded by vectors NT6, 21, 22, 23, 24, 25 and 27.
In one embodiment, the antigen receptor comprises only 1 copy of the cytoplasmic domain of human 4-1BB, 1 copy of the cytoplasmic domain of human CD28, and 1 copy of the cytoplasmic domain of human CD3 ζ. The antigen receptor may also comprise only 1 copy of the transmembrane domain of CD28 and 1 copy of the transmembrane domain of CD 8. In one example (as encoded by NT22), the first polypeptide chain can comprise the extracellular domain of a TCR β chain, 1 copy of the CD28 transmembrane domain, 1 copy of the cytoplasmic domain of human CD28, and 1 copy of the cytoplasmic domain of human CD3 ζ, while the second polypeptide chain can comprise the extracellular domain of a TCR α chain, 1 copy of the transmembrane domain of CD8, and 1 copy of the cytoplasmic domain of human 4-1 BB.
In one embodiment, the antigen receptor comprises 2 copies of the cytoplasmic domain of human 4-1BB, 2 copies of the cytoplasmic domain of human CD28, and 2 copies of the cytoplasmic domain of human CD3 ζ. For example, each of the two chains can comprise 1 copy of each of the cytoplasmic domain of human 4-1BB, the cytoplasmic domain of human CD28, and the cytoplasmic domain of human CD3 ζ (e.g., encoded by NT 6). Alternatively, one polypeptide chain (e.g., the first polypeptide chain) can comprise 2 copies of the cytoplasmic domain of human CD28, while the other (e.g., the second polypeptide chain) can comprise 2 copies of the cytoplasmic domain of human 4-1 BB. Examples include those encoded by vector NT 25.
In one embodiment of the antigen receptor, one of the two chains has the cytoplasmic domain of human CD3 ζ. For example, the first polypeptide chain can comprise 1 copy of the transmembrane domain of human CD28, 1 copy of the cytoplasmic domain of human CD28, and 1 copy of the cytoplasmic domain of human CD3 ζ. The second polypeptide chain can comprise 1 copy of the transmembrane domain of human CD8 and 1 copy of the cytoplasmic domain of human 4-1BB, but not the cytoplasmic domain of human CD3 ζ. Examples include those encoded by NT 22.
In one embodiment, the first polypeptide chain and the second polypeptide chain have different co-stimulatory domains. For example, the first polypeptide chain can comprise 1 copy of the cytoplasmic domain of human CD8, 1 copy of the cytoplasmic domain of human CD28, and 1 copy of the cytoplasmic domain of human CD3 ζ. The second polypeptide chain can comprise 1 copy of the transmembrane domain of human CD8, 1 copy of the cytoplasmic domain of human 4-1BB, and 1 copy of the cytoplasmic domain of human CD3 ζ. Examples include those encoded by NT 24.
In one embodiment, said first polypeptide chain comprises 1 copy of said transmembrane domain of human CD8, 2 copies of said cytoplasmic domain of human CD28, and 1 copy of said cytoplasmic domain of human CD3 ζ. Said second polypeptide chain comprises 1 copy of said transmembrane domain of human CD8, 2 copies of said cytoplasmic domain of human 4-1BB, and 1 copy of said cytoplasmic domain of human CD3 ζ. Examples include those encoded by NT 25.
Surprisingly and unexpectedly, (1) second generation (e.g., NT2, NT3, NT4 and NT5) and third generation (e.g., NT24) anti-NY-ESO-1/A2 TCR-based CARs show higher surface expression on virally transduced human T cells than native form TCRs (e.g., NT1 and NT1b) as determined by flow cytometry measurements, (2) second generation (e.g., NT2, NT3, NT4, NT5) and third generation (e.g., NT24) more effective in TCR-CAR mediated cytokine secretion (e.g., IL-2) and cell expansion following engagement of human T cells expressing native form TCRs (e.g., NT1, NT1b) anti-NY-ESO-1/A2 TCR-based CARs with tumor target cells, and (3) in vivo assays involving xenograft mouse Saos-2 tumor treatment models, the data indicate that the second generation anti-NY-ESO-1 TCR-CAR (e.g., NT2, NT4) showed better anti-tumor activity compared to the native form of NT1(NT1a), while the third generation anti-NY-ESO-1/A2 TCR-CAR (e.g., NT24) was significantly more effective than its second generation counterpart moiety (e.g., NT2, NT4) with only TCR signal from human CD28 (e.g., NT2) or from 4-1BB (e.g., NT4), and even more effective than the native form of NT1 (e.g., NT1 a).
In the antigen receptors described above, the antigen binding site may bind to a tumor antigen, or a Tumor Associated Antigen (TAA), or a viral antigen in a mhc (hla) -restricted manner. That is, when expressed on a cell, the extracellular domain binds to a tumor antigen, or TAA, or viral antigen in an mhc (hla) -restricted manner. A variety of such antigens are well known in the art and some examples are listed herein, including that the tumor antigen is NY-ESO-1.
Disclosed herein are methods of making the antigen receptors and cells expressing the same. The first and second polypeptide chains can be expressed from two separate expression cassettes or vectors or from one common expression cassette/vector by means of an Internal Ribosome Entry Site (IRES).
Both chains may be expressed as a fusion protein. Accordingly, the present disclosure also provides a fusion protein comprising the first polypeptide chain and the second polypeptide chain. In that case, the first polypeptide chain and the second polypeptide chain can be connected by a protein linker sequence or a self-cleaving peptide sequence. Examples of such self-cleaving peptide sequences include P2A, E2A, F2A, or T2A sequences.
The present disclosure also provides an isolated nucleic acid or a set of isolated nucleic acids encoding an antigen receptor or fusion protein as described above. In one embodiment, the disclosure also provides an isolated nucleic acid or a set of isolated nucleic acids that can comprise (I) a first nucleic acid sequence encoding the first polypeptide chain comprising a first polypeptide segment of at least 10 (e.g., 10, 20, 30, 40, 50, 60, 70, 80, 100, 150, 200, 250, and 300) amino acids (aa.) in length, and (II) a second nucleic acid sequence encoding the second polypeptide chain comprising a second polypeptide segment of at least 10 (e.g., 10, 20, 30, 40, 50, 60, 70, 80, 100, 150, 200, 250, and 300) amino acids in length. The first polypeptide segment is at least 90% (e.g., 95%, 96%, 97%, 98%, 99% or 100%) identical to the second polypeptide segment, and the first and second nucleic acid sequences comprise at least one different codon within a codon that encodes the same amino acid residue in the first and second polypeptide segments. Said first polypeptide chain and said second polypeptide chain comprise the same polypeptide sequence of greater than 10 amino acids in length; and said first nucleic acid sequence and said second nucleic acid sequence comprise at least one different codon within said codon encoding the same polypeptide sequence of said first nucleic acid sequence and said second nucleic acid sequence. In some embodiments, at least 2% (e.g., 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, or 100%) of the codons are different. The first nucleic acid sequence and the second nucleic acid sequence are less than 98% (e.g., 95%, 90%, 80%, 70%, 60%, 50%, 40%, 30%, 20%, 10%, 5%, or 0%) identical within the codon.
In some embodiments, the first nucleic acid sequence and the second nucleic acid sequence contain at least one different codon within the codon encoding one or more of: (a) the TCR β chain or antigen-binding fragment thereof, (b) the TCR α chain or antigen-binding fragment thereof, (c) the transmembrane domain of CD8, (d) the transmembrane domain of CD28, (e) the cytoplasmic domain of CD3Z or fragment thereof, (f) the cytoplasmic domain of CD3E or fragment thereof, (g) the cytoplasmic domain of CD28 or fragment thereof, and (h) the cytoplasmic domain of 4-BB or fragment thereof. The isolated nucleic acid may encode one selected from the group consisting of: SEQ ID NO: 9. 64, 10, 11 and 32-41. The isolated nucleic acid may comprise a sequence selected from the group consisting of: SEQ ID NO: 12. 65, 13, 14 and 54-63.
The nucleic acids described above may be used to express the antigen receptors described herein. Accordingly, the present disclosure also includes a vector comprising the isolated nucleic acid described above. In some embodiments, the vector is an expression vector comprising a viral vector as described herein. When expressing the first polypeptide chain and the second polypeptide chain in a cell, a bicistronic or polycistronic expression vector can be used. IRES has been widely used in various strategies for constructing bicistronic or polycistronic positive vectors. Self-cleaving 2A peptides may also be good candidates for use alone or with IRES. Thus, the first nucleic acid sequence and the second nucleic acid sequence may be linked by a nucleic acid sequence comprising an IRES. The present disclosure also provides a cell comprising an antigen receptor, fusion protein, isolated nucleic acid, or vector as described above. Examples of cells are described below, including lymphocytes, such as T cells.
Also provided are pharmaceutical compositions comprising (i) a nucleic acid, vector or cell as described above, and (ii) a pharmaceutically acceptable carrier. Such pharmaceutical compositions may be used in methods of treating neoplastic or viral infectious diseases. The method comprises administering to a subject in need thereof an effective amount of the pharmaceutical composition.
The details of one or more embodiments of the invention are set forth in the description below. Other features, objects, and advantages of the invention will be apparent from the description and from the claims.
Drawings
Fig. 1A and 1B are a set of diagrams showing exemplary antigen receptors. (A) Schematic representation of four anti-NY-ESO-1 TCR or TCR-based CAR constructs: 1(NT 1); 2(NT1 b); 3(NT 2); and 4(NT 3). (B) Schematic representation of another 11 anti-NY-ESO-1 TCR-based CAR constructs: 5(NT 4); 6(NT 5); 7(NT 6); 8(NT 21); 9(NT 22); 10(NT 23); 11(NT 24); 12(NT 25); 13(NT 26); 14(NT 27); and 15(NT 28).
FIG. 2A, FIG. 2B, FIG. 2C and FIG. 2D are schematic representations of four nucleotide sequences encoding anti-NY-ESO-1 TCR or TCR-based CAR constructs. A: NT1(SEQ ID NO: 12); b: NT1b (SEQ ID NO: 65); c: NT2(SEQ ID NO: 13); d: NT3(SEQ ID NO: 14).
FIG. 3A shows the amino acid sequence of anti-NY-ESO-1 TCR alpha chain-2A-TCR beta chain (NT 1; aNY-TCRa/b) (SEQ ID NO: 9).
FIG. 3B shows the amino acid sequence of anti-NY-ESO-1 TCR beta chain-2A-TCR alpha chain (NT 1B; aNY-TCRb/a) (SEQ ID NO: 64).
FIG. 4 shows the amino acid sequence of NT 2-anti-NY-ESO-1. beta. CD28 TmCytZCyt-2A-. alpha. mucD28TmCytmuZCyt (aNY-TCR28Z) (SEQ ID NO: 10).
FIG. 5 shows the amino acid sequence of anti-NY-ESO-1 β CD28TmCytECyt-2A- α muCD28TmCytmuECyt (NT 3; aNY-TCR28E) (SEQ ID NO: 11).
FIG. 6 shows an alignment of the nucleotide sequences hCD28TmCyt (SEQ ID NO:17) and MuhCD28TmCy (SEQ ID NO: 18). The two DNA sequences encode the same polypeptide sequence comprising the Tm and Cyt domains of native human CD28 (CD28TmCyt) (SEQ ID NO: 6). The overall homology between these two sequences is 56%. Identical nucleotides are indicated by an asterisk.
FIG. 7 shows an alignment of the nucleotide sequences hCD3ZCyt (ZCyt) (SEQ ID NO:19) and muhCD3ZCyt (muZCyt) (SEQ ID NO: 20). These two DNA sequences encode the same polypeptide sequence, which comprises the Cyt domain of native human CD3Z (SEQ ID NO: 7). The overall homology between these two sequences is 57%. Identical nucleotides are indicated by an asterisk.
FIG. 8 shows an alignment of the nucleotide sequences hCD28TmCytCD3ZCyt (28TmCytZCyt) (SEQ ID NO:16) and muhCD28TmCytCD3ZCyt (mu28TmCytZCyt) (SEQ ID NO: 21). The two DNA sequences encode the same polypeptide sequence comprising the Tm and Cyt domains of native human CD28 and Cyt of native human CD3Z (SEQ ID NO: 15). The overall homology between these two sequences is 59%. Identical nucleotides are indicated by an asterisk.
FIG. 9 shows an alignment of the nucleotide sequences hCD3ECyt (ECyt) (SEQ ID NO:23) and muhCD3ECyt (muECyt) (SEQ ID NO:24) containing a single silent nucleotide mutation (G to C) at position 119 of human CD3E (Genbank accession No.: NM-000733.1) (SEQ ID NO:23) to disrupt the BspE 1 site. These two DNA sequences encode the same polypeptide sequence comprising the Cyt domain of native human CD3E chain (CD3E) (ECyt) (SEQ ID NO: 8). The overall homology between these two sequences is 57%. Identical nucleotides are indicated by an asterisk.
FIG. 10 shows an alignment of the nucleotide sequences of Cyt of CD3Z and the mutant Cyt of CD 3Z. The CD3ZCyt sequence (SEQ ID NO:19) is from human CD3Z (GenBank ID: J04132.1). The sequences of hCD3ZCyte and mu2hCD3ZCyt (SEQ ID NO:20) encode the same amino acid sequence (SEQ ID NO: 7). The vertical bar "|" indicates the same nucleotide between the two sequences. The overall homology between these two sequences is 60.42%.
FIG. 11 shows an alignment of the nucleotide sequences of the Tm of CD28 and the mutant Tm of CD 28. The CD28Tm sequence (SEQ ID NO:44) was from human CD28(GenBank ID: BC 093698.1). The sequences of CD28Tm and mu2CD28Tm (SEQ ID NO:45) encode the same amino acid sequence (SEQ ID NO: 27). The vertical bar "|" indicates the same nucleotide between the two sequences. The overall homology between these two sequences was 62.96%.
Fig. 12 shows an alignment of the nucleotide sequences of Cyt of CD28 and mutant Cyt of CD 28. The CD28Cyt sequence (SEQ ID NO:46) is from human CD28(GenBank ID: BC 093698.1). The sequences of hCD28Cyte and mu2hCD28Cyt (SEQ ID NO:47) encode the same amino acid sequence (SEQ ID NO: 28). The vertical bar "|" indicates the same nucleotide between the two sequences. The overall homology between these two sequences is 53.65%.
FIG. 13 shows an alignment of the nucleotide sequences of Cyt of 4-1BB and the mutant Cyt of 4-1 BB. The BBCyt sequence (SEQ ID NO:48) is from human 4-1BB (GenBank ID: U03397.1). The sequences of hBBCyt and muhBBCyt (SEQ ID NO:49) encode the same amino acid sequence (SEQ ID NO: 29). The vertical bar "|" indicates the same nucleotide between the two sequences. The overall homology between these two sequences was 63.49%.
FIG. 14 shows an alignment of the nucleotide sequences of the hCD8 hinge (SEQ ID NO:50) and the MuhCD8 hinge (SEQ ID NO: 51). These two DNA sequences encode the same polypeptide sequence comprising the hinge domain of native human CD8 (CD8 hinge) (SEQ ID NO:30) (amino acid sequence 135-180 of human CD8A (GenBank ID: NM-001768.4)). The overall homology between these two sequences was 58.70%. The same nucleotides are indicated with vertical lines "|".
FIG. 15 shows an alignment of the nucleotide sequences of hCD8Tm (SEQ ID NO:52) and MuhCD8Tm (SEQ ID NO: 53). The two DNA sequences encode the same polypeptide sequence comprising the Tm domain of native human CD8 (CD8Tm) (SEQ ID NO:31) (amino acid sequence 181-206 of human CD8A, GenBank ID: NM-001768.4). The overall homology between these two sequences was 62.82%. The same nucleotides are indicated with vertical lines "|".
FIG. 16 shows the sequence of NT 4-anti-NY-ESO-1. beta. CD8 tmBBCytZCyt-2A-. alpha. mucD8TmBBCytmuZCyt (aNY-TCRBBZ) (SEQ ID NO: 32).
FIG. 17 shows the sequence of NT 5-anti-NY-ESO-1. beta. CD28 tmBBCytZCyt-2A-. alpha. mucD28TmBBCytmuZCyt (aNY-TCRBBZ) (SEQ ID NO: 33).
FIG. 18 shows the sequence of NT 6-anti-NY-ESO-1. beta. CD28 TmCytZCyt-2A-. alpha. mucD28TmCytmuZCyt (aNY-TCR28BBZ) (SEQ ID NO: 34).
FIG. 19 shows the amino acid sequence of NT21(SEQ ID NO: 35).
FIG. 20 shows the amino acid sequence of NT22(SEQ ID NO: 36).
FIG. 21 shows the amino acid sequence of NT23(SEQ ID NO: 37).
FIG. 22 shows the amino acid sequence of NT24(SEQ ID NO: 38).
FIG. 23 shows the amino acid sequence of NT25(SEQ ID NO: 39).
FIG. 24 shows the amino acid sequence of NT26(SEQ ID NO: 40).
FIG. 25 shows the amino acid sequence of NT27(SEQ ID NO: 41).
FIGS. 26A and 26B are graphs showing surface expression of anti-NY-ESO-1 TCR-CAR on infected PG13 VPC. PG13 was infected with anti-NY-ESO-1 TCR-CAR + (NT22 (vector 9; third generation), NT24 (vector 11; third generation), NT25 (vector 12; second generation)). (A) The intensity plots show positive staining for FTIC-anti-human TCRVb13.1 and APC-NYPep/A2 tetramer single or double positive cells, respectively. The% of positive cells are shown in a single quadrant. (B) The histogram shows the% of cells staining positive for the NYPep/A2 tetramer. Untd: is not infected.
FIGS. 27A and 28B are graphs showing surface expression of the native form of anti-NY-ESO-1 TCR or second generation TCR-CAR on transduced activated human T cells. The ATC evaluated was transduced with one of two anti-NY ESO-1 TCRs (NT1, NT1b) or one of two second generation TCR-based CARs (NT2 and NT 3). (A) The intensity plots show positive staining for FTIC-anti-human TCRVb13.1 and APC-NYPep/A2 tetramer single or double positive cells, respectively. The percentage of positive cells is shown in a single quadrant. (B) The histogram shows the percentage of cells staining positive for the NYPep/A2 tetramer.
FIG. 28 is a set of photographs showing the specific killing effect on tumor cells (Saos-2) using ATC transduced with one of two anti-NY ESO-1 TCRs (NT1, NT1b) or one of two second generation TCR-based CARs (NT2 and NT 3).
FIGS. 29A and 29B are graphs showing cytokine secretion from ATC transduced with one of two anti-NY ESO-1 TCRs (NT1, NT1B) or one of two second generation TCR-based CARs (NT2 and NT 3). (A) IL-2 secretion and (B) INF- γ secretion, with data expressed as mean ± s.d. from three samples in each group.
FIG. 30 is a graph showing the change in the percentage of NYPep/A2+ cells of ATC transduced with one of two anti-NY ESO-1 TCRs (NT1, NT1b) or one of two second generation TCR-based CARs (NT2 and NT3) following co-culture with tumor target cells (Saos-2).
FIGS. 31A and 31B are graphs showing surface expression of the native form of anti-NY-ESO-1 TCR or second or 3 rd generation TCR-CAR on transduced activated human T cells. ATC were evaluated as transduced with or untransduced with NY-ESO-1 TCR-resistant (NT1 (also referred to as NT1 a); vector 1) or NY-ESO-1TCR-CAR + (NT2 (vector 3; second generation), NT4 (vector 5; second generation), NT5 (vector 6; second generation), NT24 (vector 11; third generation)). (A) The intensity plots show positive staining for FITC-anti-human TCRVb13.1 and APC-NYPep/A2 tetramer single or double positive cells, respectively. The% of positive cells are shown in a single quadrant. (B) The histogram shows cells staining positive for the NYPep/A2 tetramer (numbers in parentheses indicate geometric mean).
FIG. 32 is a graph showing target cell killing effect of human ATC transduced with anti-NY ESO-1TCR or one of the four TCR-based CARs after conjugation to target cells (Saos-2). ATC were evaluated as transduced with or untransduced with NY-ESO-1 TCR-resistant (NT1 (also referred to as NT1 a); vector 1) or NY-ESO-1TCR-CAR + (NT2 (vector 3; second generation), NT4 (vector 5; second generation), NT5 (vector 6; second generation), NT24 (vector 11; third generation)).
FIGS. 33A and 33B are graphs showing (A) IL-2 secretion and (B) IFN- γ of human ATC transduced with anti-NY ESO-1TCR or one of the four TCR-based CARs following conjugation to target cells (Saos-2). ATC was evaluated as transduced with anti-NY-ESO-1 TCR (NT1 (also referred to as NT1 a); vector 1) or anti-NY-ESO-1 TCR-CAR + (NT2 (vector 3; second generation), NT4 (vector 5; second generation), NT5 (vector 6; second generation), NT24 (vector 11; third generation)). ATC is transduced with anti-NY-ESO-1 TCR (NT1 (also known as NT1 a); vector 1) or anti-NY-ESO-1 TCR-CAR + (NT2 (vector 3; second generation), NT4 (vector 5; second generation), NT5 (vector 6; second generation), NT24 (vector 11; third generation)), or untransduced (Untd).
FIGS. 34A and 34B are graphs showing the change in (A) percentage and (B) amplification of NYPep/A2+ ATC transduced with anti-NY ESO-1TCR or one of the four TCR-based CARs following conjugation to target cells (Saos-2). ATC were evaluated as transduced with or untransduced with NY-ESO-1 TCR-resistant (NT1 (also referred to as NT1 a); vector 1) or NY-ESO-1TCR-CAR + (NT2 (vector 3; second generation), NT4 (vector 5; second generation), NT5 (vector 6; second generation), NT24 (vector 11; third generation)).
FIGS. 35A and 35B are graphs showing the in vivo therapeutic effect of second or third generation TCR-CAR expression likely to enhance anti-NY-ESO-1/A2 TCR CAR mediated anti-tumor activity in human T cells in a xenograft Saos-2 tumor mouse model. ATC evaluated in vivo antitumor therapy experiments were transduced with anti-NY ESO-1TCR (NT1 (also known as NT1 a); vector 1) or anti-NY-ESO-1 TCR-CAR + (NT2 (vector 3; second generation), NT4 (vector 5; second generation), NT5 (vector 6; second generation), NT24 (vector 11; third generation)), or untransduced (Untd). (A) Mean tumor size + s.d. from 6 mice in each group. (B) The percent survival of each group of mice was calculated and given.
Detailed Description
The present disclosure relates to agents, methods, and compositions that confer and/or enhance an immune response mediated by cellular immunotherapy (such as by adoptively transferring subsets of lymphocytes that are genetically modified with antigen-specificity).
Such adoptive cell transfer or Adoptive Cell Therapy (ACT) represents a promising therapeutic approach for treating cancer patients. However, it faces various obstacles, such as loss of stability for efficient CAR expression and activity. The present disclosure addresses such obstacles in a number of ways, including but not limited to (1) novel designs of third generation TCR-CARs that introduce co-stimulatory signaling, e.g., human 4-1BB, (2) novel designs involving variation in different transmembrane domains, different copy numbers of the same TCR signaling element (e.g., CD3Z and co-stimulatory molecules of CD28 or 4-1 BB). The methods described herein enhance TCR-CAR mediated T cell signaling by genetically expressing a novel third generation TCR-CAR in T cells to improve the anti-tumor capacity of TCR-CAR expressing T cells.
The present disclosure provides compositions comprising genetically modified lymphocytes expressing chimeric antigen receptors having the ability to modulate the immune system as well as innate and adaptive immune responses. The disclosed agents, methods, and compositions provide genetically engineered lymphocytes with enhanced anti-tumor function, as well as methods of developing such lymphocytes.
Antigen receptor
Genetically modified immune functional cells (e.g., T cells and NK cells) engineered to express foreign antigen receptors are effective immunotherapies for cancer and infectious diseases. The isolation of autoantigen-specific immune cells (e.g., T cells) for therapeutic applications is a difficult task and is not possible in cases where such cells are absent or rare. Thus, strategies have been developed to genetically transfer immune receptors specific for tumors or viruses to patient T cells. To this end, antigen receptors have been constructed that link an antigen (Ag) -recognition domain to the signaling domain of a TCR or Fc receptor. T cells expressing this antigen receptor reproduce an immune specific response mediated by the introduced receptor.
Chimeric antigen receptors (also known as chimeric immunoreceptors, chimeric T cell receptors, or artificial T cell receptors) are receptor proteins that have been engineered to give T cells a new ability to target specific proteins. The receptor is chimeric in that it binds both antigen binding and T cell activation functions into one receptor. In addition to the antigen binding site, the CAR has one or more functional domains.
Domain
As described herein, an antigen receptor comprises three domains: the extracellular domain, transmembrane domain, and cytoplasmic domain (which may contain an intracellular signaling domain). In some embodiments, it contains a fourth domain: an extracellular hinge region between the extracellular and cytoplasmic domains. Thus, the chimeric antigen receptor combines many aspects of normal T cell activation into a single protein. It links the extracellular antigen recognition domain to an intracellular signaling domain which, upon antigen binding, activates T cells.
The extracellular domain comprises an antigen-binding or target-binding domain. Exposed to the outside of the cell, this domain interacts with potential target molecules and is responsible for targeting CAR-T cells to any cell expressing the matching molecule. The antigen recognition domain may typically be derived from the variable regions of monoclonal antibodies linked together as single chain variable fragments (scfvs) or from TCRs. In addition to antibodies and TCRs, other methods can be used to direct the specificity of the CAR, typically utilizing ligand/receptor pairs that typically bind to each other. For example, cytokines, innate immunity receptors, TNF receptors, growth factors and structural proteins may be used as antigen recognition domains. In preferred embodiments, the extracellular domain comprises the variable region of an antibody or a functional fragment thereof, the extracellular domain of a T cell (e.g., Va or Vb; VaCa or VbCb) or a functional fragment thereof.
The hinge region (also referred to as a spacer) is a small domain located between the antigen recognition region and the outer membrane of the cell. The ideal hinge region can enhance the flexibility of the receptor's target binding domain, thereby reducing the spatial constraint between the CAR and its target antigen. This promotes antigen binding and synapse formation between the CAR-T cell and the target cell. The hinge sequence may comprise the membrane proximal region from an immune molecule such as IgG, CD8 and CD 28.
The transmembrane domain is a structural component, which consists of a hydrophobic alpha helix spanning the cell membrane. It anchors the antigen receptor to the plasma membrane, bridging the extracellular hinge and the antigen recognition domain with cytoplasmic/intracellular signaling regions. This domain is important for the stability of the entire receptor. In the present disclosure, transmembrane domains from the membrane proximal component of the cytoplasm may be used, but different transmembrane domains may lead to different receptor stabilities. As used herein, a transmembrane domain may comprise a functional transmembrane domain known in the art (e.g., a transmembrane domain of CD3 ζ, CD28, CD8, CD4, or fcepsilon Ri γ, or a variant polypeptide or functional fragment thereof). The CD28 transmembrane domain is known to result in highly expressed stable receptors.
Intracellular T cell signaling domains are located in the cytoplasmic domain of receptors inside the cell. Upon binding of the antigen to the external antigen recognition domain, the receptors aggregate together and transmit an activation signal. The internal cytoplasmic end of the receptor then permanently transmits the signal inside the T cell. Normal T cell activation is dependent on phosphorylation of immunoreceptor-based tyrosine activation motifs (ITAMs) present in the cytoplasmic domain of CD 3-zeta. Thus, the cytoplasmic domain of CD 3-zeta can be used. Other ITAM-containing domains may also be used. For example, an intracellular signaling domain can comprise, e.g., a cytoplasmic portion of CD3 ζ or functional fragment thereof, FC epsilon RI γ or functional fragment thereof, and/or CD28 or functional fragment thereof.
In addition to CD3 signaling, T cells also use co-stimulatory molecules so as to persist after activation. Thus, the cytoplasmic domain of the CAR receptor may also comprise one or more chimeric domains from a costimulatory protein (i.e., a costimulatory domain). Signaling domains from a variety of co-stimulatory molecules may be used. Examples of co-stimulatory polypeptides known to stimulate or increase an immune response by their binding include CD28, OX-40, 4-1BB, CD27, and NKG2D and their corresponding ligands, including B7-1, B7-2, OX-40L, 4-1BBL, CD70, and NKG2D ligands. Such polypeptides are present in the tumor microenvironment and activate an immune response to neoplastic cells. In various embodiments, promotion, stimulation, or agonism of the proinflammatory polypeptide and/or its ligand by the therapeutic transgene can enhance the immune response of the immune responsive cell. For example, co-stimulation of CD28 (signal 2) during T cell activation by TCR (signal 1) leads to sustained proliferation, activation-induced reduction of cell death (AICD), and improvement in long-term lymphocyte survival. Examples of domain sequences that may be included in a CAR are listed in the table below.
TABLE A
The two major antigen receptors used to re-target T cells are the transgenic T cell receptor (tcr) and the Chimeric Antigen Receptor (CAR). CARs can be divided into two forms, antibody-based CARs and TCR-based CARs. Antibody-based CAR therapies have demonstrated great success in targeting B-cell leukemia, and trials against solid tumors are ongoing. Although antibody-based CARs have great potential as therapeutic agents in cancer immunotherapy, they are limited in their ability to recognize only cell surface molecules. In contrast, tTCR and TCR-based CARs have the ability to recognize any processed antigen presented by the Major Histocompatibility Complex (MHC), greatly expanding the range of possible targets. For example, in vitro studies have shown that cells engineered with an endogenously occurring NY-ESO-1TCR are active against melanoma and non-melanoma cell lines expressing NY-ESO-1. In a recent clinical trial using NY-ESO-1 targeted T cells, engineered cells carrying high affinity tTCR were delivered to patients with melanoma and synovial cell carcinoma. Nearly half of the patients in this study showed objective clinical remission, highlighting the potential of tcr T cells in the treatment of established solid tumors.
There are two ways to genetically introduce TCR-associated antigen-specific genes into T cells: t cells are genetically engineered to express exogenous native TCRs (e.g., TCR α and TCR β chains) and T cells are genetically engineered to express exogenous TCR-based CARs. TCR-based CARs can be established by: comprising an extracellular domain (e.g., of a TCR α chain or a TCR β chain), a transmembrane domain, and a TCR signaling element (e.g., the cytoplasmic domain of a CD3 zeta chain (CD3Z) or CD3E chain (CDE 3)), with or without integration of a costimulatory signaling element, such as the cytoplasmic domain of CD28 or CD137(4-1 BB). One advantage of TCR-based CARs, as compared to native TCRs, is that they can comprise TCR signaling elements, such as CD3Z and CDE, and be integrated with co-stimulatory signaling elements, such as CD28 and CD137(4-1 BB).
It has been found that co-stimulation of CD28 (signal 2) during T cell activation by TCRs (signal 1; e.g., CD 3Z-or CD 3E-mediated signaling) can promote continued T cell proliferation. To combine activation and co-stimulatory functions in a single receptor, CARs consisting of CD3Z and CD28 sequences in the same molecule can be constructed. Such IgCD28Z molecules have been demonstrated to have excellent cytotoxicity, proliferation, and functions of IL2 and IFN γ production in T cells.
The extracellular portion of the antibody-based CAR (antibody-based CAR; sFv-CAR) consists of a single-chain Fv or a fragment thereof. Like sFv-CARs, TCR-CARs can comprise TCR variable antigen-binding fragments (va and ν β, or antigen-binding fragments thereof) linked to the signaling domain of a TCR or Fc receptor in the context of specific mhc (hla) molecules. TCR-CARs can be constructed in two basic forms: a single chain TCR-CAR (scTCR-CAR) or a double chain TCR-CAR (tcTCR-CAR). For scTCR-CAR, both TCR V α and V β are present in the same single-chain TCR protein, whereas in tcTCR-CAR, V α and V β are in separate chains forming a heterodimer.
It is well known that TCRs have approximately 100 to 1,000 fold lower binding affinity for their cognate antigen (peptide/MHC complex) as compared to antibodies directed against the same cognate antigen. Furthermore, it has been reported that the single-chain form of T cell receptors generally has a greatly reduced affinity for their cognate antigen compared to their affinity for the corresponding maternal double-chain form of such antigens, with some single-chain T cell receptors losing antigen binding affinity altogether. With respect to the function of such cells in T cell activation, including induction of proliferation, killing of target cells, and induction of cytokine secretion, it has been demonstrated that higher affinity of TCRs, including chimeric TCRs, correlates with higher potency of modified T cells expressing such receptors. Several reports indicate that the binding affinity of the single chain form of TCR-CAR is too low to be of therapeutic or diagnostic value. Thus, in order to generate a functional TCR-CAR with a relatively high antigen binding affinity, the double-stranded form of native TCR has a more significant advantage than the single-stranded form.
Recombination between nucleic acids is a well-recognized phenomenon in molecular biology. Genetic recombination, which requires strong sequence homology between the participating nucleic acid sequences, is generally referred to as homologous recombination. Although most gene knockout strategies employ homologous recombination to achieve targeted knockouts, in some systems, the occurrence of gene recombination can adversely affect gene manipulation. In particular, homologous recombination events can adversely affect the construction and production of vectors, particularly viral vectors (e.g., adenoviruses, retroviruses, adeno-associated viruses, herpes viruses, and the like), where it is often desirable to maintain highly homologous sequences (e.g., identical polypeptide sequences) within a single, stable viral vector without homologous recombination during, for example, passage and/or propagation of the viral vector through one or more host cells and/or organisms.
As a means of reducing possible viral recombination events, a two-vector approach can be employed to transduce two protein molecules comprising highly homologous polypeptide sequences encoded by similar highly homologous nucleic acid sequences into a single cell. For example, a TCR-CAR in a double-stranded form, which consists of various vacas and VbCb, respectively, but share the same signaling element polypeptide sequence (e.g., cytoplasmic domains of CD28 and/or CD 3Z). Two different vectors, each encoding one of the two homologous proteins of interest, can be produced from a single VPC. However, the rate of successful transduction of a single vector into mammalian cells (e.g., activated T cells exposed to a retrovirus) is often limited.
A new approach to overcome such problems associated with low host cell transduction efficiency is to enable the delivery of two or more nucleic acid sequences encoding highly homologous (e.g., identical) polypeptides on a single viral vector. This approach allows one to produce viral vector sequences comprising nucleic acid sequences encoding two or more highly homologous (e.g., identical) polypeptides or polypeptide domains thereof. The viral vector sequences have a reduced risk of e.g. homologous recombination between nucleic acid sequences even during prolonged passages and/or multiple infection, chromosomal integration and/or excision events, e.g. in a host cell. Such reduced homologous recombination rates are at least partially due to the development of degeneracy in the genetic code during viral vector synthesis as described herein.
In addition, the hinge/spacer consists of the non-antigen binding extracellular region of the CAR. The hinge/spacer modulates CAR function by providing flexibility, lengthening length, allowing dimerization to occur, or improving stability. These properties have been shown to affect the interaction of effector cells with target cells, thereby affecting the intensity of the activation signal. Conventional hinge/spacers were prepared using immunoglobulin Fc, CD8 α, and CD28 spacers.
Examples of domain sequences and related nucleic acid sequences that may be included in the CARs disclosed herein are listed below.
1, SEQ ID NO: 1G 4a 95LY TCR alpha chain (275aa)
METLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSCDVKLVEKSFETDTNLNFQNLSVIGFRILLLKVAGFNLLMTLRLWSSR
2, SEQ ID NO: 1G 4a 95LY TCR beta chain (311aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADCGFTSESYQQGVLSATILYEILLGKATLYAVLVSALVLMAMVKRKDSRG
SEQ ID NO:3:P2A(19aa)
ATNFSLLKQAGDVEENPGP
4, SEQ ID NO: ec domain of the alpha chain of 1G 4a 95LY TCR (227aa)
METLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESS
5, SEQ ID NO: ec domain of beta chain of 1G 4a 95LY TCR (262aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRAD
6 of SEQ ID NO: human CD28TmCyt (68aa)
FWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRS
7, SEQ ID NO: human CD3ZCyt (112aa)
RVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
8, SEQ ID NO: human CD3ECyt (55aa)
KNRKAKAKPVTRGAGAGGRQRGQNKERPPPVPNPDYEPIRKGQRDLYSGLNQRRI
SEQ ID NO:9:NT1_aNY-TCR(612aa)
METLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSCDVKLVEKSFETDTNLNFQNLSVIGFRILLLKVAGFNLLMTLRLWSSRAKRSGSGATNFSLLKQAGDVEENPGPMSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADCGFTSESYQQGVLSATILYEILLGKATLYAVLVSALVLMAMVKRKDSRG
SEQ ID NO:64:NT1b_Xho_NY-ESO.txt.xprt(612aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADCGFTSESYQQGVLSATILYEILLGKATLYAVLVSALVLMAMVKRKDSRGAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSCDVKLVEKSFETDTNLNFQNLSVIGFRILLLKVAGFNLLMTLRLWSSR
SEQ ID NO:10:NT2_aNY-TCR28Z(NYESO1-TCRba28Z_Xho-NY-ESO1
bmu2CD28tmcytCD3Zcyt-P2A-1g4TCRa95LYCD28tmcytCD3Zcyt),883aa
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADGSPKFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSGSPKFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
SEQ ID NO:11:NT3_aNY-TCR28E
(NYESO1-TCRba28E_NY-ESO1bmu2CD28tmcytCD3Ecyt-P2A-1g4TCRa95LYCD28tmcytCD3Ecyt)(769aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADGSPKFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSKNRKAKAKPVTRGAGAGGRQRGQNKERPPPVPNPDYEPIRKGQRDLYSGLNQRRIAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSGSPKFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSKNRKAKAKPVTRGAGAGGRQRGQNKERPPPVPNPDYEPIRKGQRDLYSGLNQRRI
12, SEQ ID NO: encoding NT1_ aNY-TCR
(NYESO1-TCRab _ Xho-NY-ESO11g4TCRa95LY-P2A-b-Not) nucleotide sequence
ATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCTGTGATGTCAAGCTGGTCGAGAAAAGCTTTGAAACAGATACGAACCTAAACTTTCAAAACCTGTCAGTGATTGGGTTCCGAATCCTCCTCCTGAAAGTGGCCGGGTTTAATCTGCTCATGACGCTGCGGCTGTGGTCCAGCCGGGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACTGTGGCTTCACCTCCGAGTCTTACCAGCAAGGGGTCCTGTCTGCCACCATCCTCTATGAGATCTTGCTAGGGAAGGCCACCTTGTATGCCGTGCTGGTCAGTGCCCTCGTGCTGATGGCTATGGTCAAGAGAAAGGATTCCAGAGGCTAA
SEQ ID NO:65:NT1b_Xho_NY-ESO1-TCRb-P2A-1g4TCRa95LY-not.txt.xdna
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACTGTGGCTTCACCTCCGAGTCTTACCAGCAAGGGGTCCTGTCTGCCACCATCCTCTATGAGATCTTGCTAGGGAAGGCCACCTTGTATGCCGTGCTGGTCAGTGCCCTCGTGCTGATGGCTATGGTCAAGAGAAAGGATTCCAGAGGCGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCTGTGATGTCAAGCTGGTCGAGAAAAGCTTTGAAACAGATACGAACCTAAACTTTCAAAACCTGTCAGTGATTGGGTTCCGAATCCTCCTCCTGAAAGTGGCCGGGTTTAATCTGCTCATGACGCTGCGGCTGTGGTCCAGCCGGTAA
13 in SEQ ID NO: nucleotide sequence encoding NT2_ aNY-TCR28Z NYESO1-TCRba28Z _ Xho-NY-ESO1bmu2CD28tmcytCD3Zcyt-P2A-1g4TCRa95LYCD28tmcytCD3Zcyt-Not
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACGGCTCTCCTAAGTTCTGGGTTCTCGTCGTCGTGGGAGGTGTGTTAGCATGTTACTCTCTCTTGGTTACTGTCGCTTTCATAATCTTTTGGGTCCGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGTCGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGAGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCGGCTCCCCAAAATTTTGGGTGCTGGTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATTTTCTGGGTGAGGAGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCCCGCCGCCCCGGGCCCACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCCTATCGCTCCAGAGTGAAGTTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCCCCTCGCTAA
14, SEQ ID NO: encoding NT3_ aNY-TCR28E
NYESO1-TCRba28E_Xho-NY-ESO1
bmu2 nucleotide sequence of CD28tmcytCD3Ecyt-P2A-1g4TCRa95LYCD28tmcytCD3Ecyt-Not
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACGGCTCTCCTAAGTTCTGGGTTCTCGTCGTCGTGGGAGGTGTGTTAGCATGTTACTCTCTCTTGGTTACTGTCGCTTTCATAATCTTTTGGGTCCGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGTAAAAACCGCAAAGCTAAAGCTAAACCCGTCACTAGGGGGGCCGGAGCAGGAGGGCGCCAGCGCGGTCAGAATAAAGAACGCCCTCCTCCCGTCCCTAATCCTGATTACGAACCGATTAGAAAGGGGCAAAGAGATCTCTACAGCGGACTCAACCAACGGAGAATTGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCGGCTCCCCAAAATTTTGGGTGCTGGTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATTTTCTGGGTGAGGAGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCCCGCCGCCCCGGGCCCACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCCTATCGCTCCAAGAATAGAAAGGCCAAGGCCAAGCCTGTGACACGAGGAGCGGGTGCTGGCGGCAGGCAAAGGGGACAAAACAAGGAGAGGCCACCACCTGTTCCCAACCCAGACTATGAGCCCATCCGCAAAGGCCAGCGGGACCTGTATTCTGGCCTGAATCAGAGACGCATCTAA
SEQ ID NO:15:hCD28TmCytCD3ZCyt(141aa)
MNMTPRRPGPTRKHYQPYAPPRDFAAYRSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
16 in SEQ ID NO: nucleotide sequence (540bp) for coding hCD28TmCytCD3ZCyt
TTTTGGGTGCTGGTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATTTTCTGGGTGAGGAGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCCCGCCGCCCCGGGCCCACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCCTATCGCTCCAGAGTGAAGTTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCCCCTCGC
17 in SEQ ID NO: nucleotide sequence (204bp) encoding hCD28TmCyt
TTTTGGGTGCTGGTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATTTTCTGGGTGAGGAGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCCCGCCGCCCCGGGCCCACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCCTATCGCTCC
18, SEQ ID NO: nucleotide sequence (mu28TmCyt) (204bp) encoding a mutation of hCD28TmCytCD3ZCyt
TTCTGGGTTCTCGTCGTCGTGGGAGGTGTGTTAGCATGTTACTCTCTCTTGGTTACTGTCGCTTTCATAATCTTTTGGGTCCGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGT
19, SEQ ID NO: nucleotide sequence 336bp for coding human CD3ZCyt (ZCyt)
AGAGTGAAGTTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCCCCTCGC
20, SEQ ID NO: nucleotide sequence encoding a mutation of human CD3ZCyt (muZCyt), muhCD3ZCyt, 336bp
CGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGA
21, SEQ ID NO: nucleotide sequence encoding a mutation of hCD28TmCytCD3ZCyt (mu28TmCytmuCyt) muhCD28TmCytmuCD3ZCyt, 540bp
TTCTGGGTTCTCGTCGTCGTGGGAGGTGTGTTAGCATGTTACTCTCTCTTGGTTACTGTCGCTTTCATAATCTTTTGGGTCCGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGTCGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGA
22, SEQ ID NO: nucleotide sequence (165bp) of human CD3ECyt _ NM _000733.1
AAGAATAGAAAGGCCAAGGCCAAGCCTGTGACACGAGGAGCGGGTGCTGGCGGCAGGCAAAGGGGACAAAACAAGGAGAGGCCACCACCTGTTCCCAACCCAGACTATGAGCCCATCCGGAAAGGCCAGCGGGACCTGTATTCTGGCCTGAATCAGAGACGCATC
23, SEQ ID NO: nucleotide sequence of human CD3ECyt _ NM _000733.1 with a single silent nucleotide mutation (G to C) at position 119 to disrupt BspE 1 position (165bp)
AAGAATAGAAAGGCCAAGGCCAAGCCTGTGACACGAGGAGCGGGTGCTGGCGGCAGGCAAAGGGGACAAAACAAGGAGAGGCCACCACCTGTTCCCAACCCAGACTATGAGCCCATCCGCAAAGGCCAGCGGGACCTGTATTCTGGCCTGAATCAGAGACGCATC
24, SEQ ID NO: nucleotide sequence of a silent mutant human CD3ECyt _ NM _000733.1, edited, 165bp
AAAAACCGCAAAGCTAAAGCTAAACCCGTCACTAGGGGGGCCGGAGCAGGAGGGCGCCAGCGCGGTCAGAATAAAGAACGCCCTCCTCCCGTCCCTAATCCTGATTACGAACCGATTAGAAAGGGGCAAAGAGATCTCTACAGCGGACTCAACCAACGGAGAATT
25 in SEQ ID NO: human NY-ESO-1 peptides corresponding to residues 157 to 165 of human NY-ESO-1 (NY-ESO-1:157-165) (9aa)
SLLMWITQC
SEQ ID NO:26:CD28pec(40aa)
KIEVMYPPPYLDNEKSNGTIIHVKGKHLCPSPLFPGPSKP
27 of SEQ ID NO: human CD28Tm
FWVLVVVGGVLACYSLLVTVAFIIFWV
28, SEQ ID NO: human CD28Cyt
RSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRS
29 in SEQ ID NO: human 4-1BBCyt
KRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCEL
30 of SEQ ID NO: CD8 hinge txt
AKPTTTPAPRPPTPAPTIASQPLSLRPEACRPAAGGAVHTRGLDFA
SEQ ID NO:31:CD8Tm_aa 181-20.txt.xprt(26aa)
CDIYIWAPLAGTCGVLLLSLVITLYC
SEQ ID NO:32:NT4_NYESO1-TCRbaBBZ_Xho-NY-ESO1 bmuCD8tmBBcyt CD3Zcyt-P2A-1g4TCRa95LYCCD8tmBBcytZcyt-Not.txt.xprt(883aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADGSPKCDIYIWAPLAGTCGVLLLSLVITLYCKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSGSPKCDIYIWAPLAGTCGVLLLSLVITLYCKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
33, SEQ ID NO: NT5_ DESIGN _ NYE.txt.xprt (885aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADGSPKFWVLVVVGGVLACYSLLVTVAFIIFWVKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSGSPKFWVLVVVGGVLACYSLLVTVAFIIFWVKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
SEQ ID NO:34:NT6_NYESO1-TCRb.txt.xprt(967 aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADGSPKFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSGSPKFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
SEQ ID NO:35:NT21_.txt.xprt(883 aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADGSPKFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSGSPKCDIYIWAPLAGTCGVLLLSLVITLYCKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
SEQ ID NO:36:NT22_.txt.xprt(771 aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADGSPKFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSGSPKCDIYIWAPLAGTCGVLLLSLVITLYCKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCEL
SEQ ID NO:37:NT23_.txt.xprt(884 aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADGSPKFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSGSPKFWVLVVVGGVLACYSLLVTVAFIIFWVKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
SEQ ID NO:38:NT24_.txt.xprt(882 aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADGSPKCDIYIWAPLAGTCGVLLLSLVITLYCRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSGSPKCDIYIWAPLAGTCGVLLLSLVITLYCKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
SEQ ID NO:39:NT25_.txt.xprt(965 aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADGSPKCDIYIWAPLAGTCGVLLLSLVITLYCRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSGSPKCDIYIWAPLAGTCGVLLLSLVITLYCKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
SEQ ID NO:40:NT26_.txt.xprt(1059 aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADGSPKAKPTTTPAPRPPTPAPTIASQPLSLRPEACRPAAGGAVHTRGLDFAPRKIEVMYPPPYLDNEKSNGTIIHVKGKHLCPSPLFPGPSKPFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSGSPKAKPTTTPAPRPPTPAPTIASQPLSLRPEACRPAAGGAVHTRGLDFAPRKIEVMYPPPYLDNEKSNGTIIHVKGKHLCPSPLFPGPSKPFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
SEQ ID NO:41:NT27_.txt.xprt(859 aa)
MSIGLLCCAALSLLWAGPVNAGVTQTPKFQVLKTGQSMTLQCAQDMNHEYMSWYRQDPGMGLRLIHYSVGAGITDQGEVPNGYNVSRSTTEDFPLRLLSAAPSQTSVYFCASSYVGNTGELFFGEGSRLTVLEDLKNVFPPEVAVFEPSEAEISHTQKATLVCLATGFYPDHVELSWWVNGKEVHSGVSTDPQPLKEQPALNDSRYALSSRLRVSATFWQDPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRADGSPKAKPTTTPAPRPPTPAPTIASQPLSLRPEACRPAAGGAVHTRGLDFAPRKIEVMYPPPYLDNEKSNGTIIHVKGKHLCPSPLFPGPSKPFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRAKRSGSGATNFSLLKQAGDVEENPGPMETLLGLLILWLQLQWVSSKQEVTQIPAALSVPEGENLVLNCSFTDSAIYNLQWFRQDPGKGLTSLLLIQSSQREQTSGRLNASLDKSSGRSTLYIAASQPGDSATYLCAVRPLYGGSYIPTFGRGTSLIVHPYIQNPDPAVYQLRDSKSSDKSVCLFTDFDSQTNVSQSKDSDVYITDKTVLDMRSMDFKSNSAVAWSNKSDFACANAFNNSIIPEDTFFPSPESSGSPKCDIYIWAPLAGTCGVLLLSLVITLYCKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCEL
42 of SEQ ID NO: nucleotide sequence (120bp) of pEc of hCD 28-human CD 28-complete CDs-mRNA-BC093698.1. txt. xdna
AAAATTGAAGTTATGTATCCTCCTCCTTACCTAGACAATGAGAAGAGCAATGGAACCATTATCCATGTGAAAGGGAAACACCTTTGTCCAAGTCCCCTATTTCCCGGACCTTCTAAGCCC
43 of SEQ ID NO: nt of mu2hCD28 pEc-human CD 28-complete CDs-mRNA-BC093698.1. txt. xdna (120bp)
AAGATCGAGGTAATGTACCCACCGCCCTATCTTGATAACGAAAAATCTAACGGTACAATAATTCACGTCAAGGGCAAGCATTTGTGCCCTTCCCCGTTGTTCCCGGGCCCAAGCAAACCG
44 of SEQ ID NO: nucleotide sequence (81bp) of CD28Tm _ human CD28_ BC093698.1.txt.xdna
TTTTGGGTGCTGGTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATTTTCTGGGTG
45 in SEQ ID NO: nucleotide sequence of mu2CD28 Tm-human CD 28-BC093698.1. txt. xdna (81bp)
TTCTGGGTTCTCGTCGTCGTGGGAGGTGTGTTAGCATGTTACTCTCTCTTGGTTACTGTCGCTTTCATAATCTTTTGGGTC
46 of SEQ ID NO: nucleotide sequence of hCD28Cyt.txt.xdna (123bp)
AGGAGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCCCGCCGCCCCGGGCCCACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCCTATCGCTCC
47 of SEQ ID NO: nucleotide sequence of mu2hCD28Cyt. txt. xdna (123bp)
CGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGT
48 of SEQ ID NO: design the nucleotide sequence of _ h4-1BBCyt. xdna (126bp)
AAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTG
49 of SEQ ID NO: design the nucleotide sequence of _ muh41BBCyt. xdna (126bp)
AAGAGAGGGCGTAAAAAGCTGCTCTACATCTTTAAGCAGCCTTTCATGCGTCCTGTTCAGACAACACAGGAAGAGGACGGATGCTCTTGCAGGTTCCCTGAGGAGGAGGAGGGTGGTTGCGAGCTC
50 of SEQ ID NO: nucleotide sequence (138bp) of CD8 hinge
GCTAAGCCCACCACGACGCCAGCGCCGCGACCACCAACACCGGCGCCCACCATCGCGTCGCAGCCCCTGTCCCTGCGCCCAGAGGCGTGCCGGCCAGCGGCGGGGGGCGCAGTGCACACGAGGGGGCTGGACTTCGCC
51 of SEQ ID NO: nucleotide sequence (138bp) of hinge of muCD8
GCTAAGCCCACTACTACCCCAGCTCCCAGGCCTCCCACACCTGCCCCAACAATCGCCAGCCAGCCACTGTCCCTTAGGCCCGAGGCCTGTAGGCCCGCCGCCGGAGGAGCCGTGCACACCCGCGGACTGGATTTTGCT
52, SEQ ID NO: h _ CD8A _ NM _001768.4. xdna's CD8Tm _ aa 181- & 206 nucleotide sequence (78bp)
TGTGATATCTACATCTGGGCGCCCTTGGCCGGGACTTGTGGGGTCCTTCTCCTGTCACTGGTTATCACCCTTTACTGC
53, SEQ ID NO: nucleotide sequence of MuCD8Tm, 78bp
MuCD8Tm_aa 181-206of H_CD8A_NM_001768.4
TGCGACATTTATATTTGGGCCCCTCTCGCTGGCACATGCGGCGTGTTGTTGCTCAGCCTCGTGATTACACTTTATTGT
54, SEQ ID NO: NT 4-design-NYESO 1-TCRbaBBZ-Xho-NY-ESO 1
Nucleotide sequence of bmuCD8tmBBcytCD3Zcyt-P2A-1g4TCRa95LYCCD8tmBBcytZcyt-not
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACGGCTCTCCTAAGTGCGACATTTATATTTGGGCCCCTCTCGCTGGCACATGCGGCGTGTTGTTGCTCAGCCTCGTGATTACACTTTATTGTAAGAGAGGGCGTAAAAAGCTGCTCTACATCTTTAAGCAGCCTTTCATGCGTCCTGTTCAGACAACACAGGAAGAGGACGGATGCTCTTGCAGGTTCCCTGAGGAGGAGGAGGGTGGTTGCGAGCTCCGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGAGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCGGCTCCCCAAAATGTGATATCTACATCTGGGCGCCCTTGGCCGGGACTTGTGGGGTCCTTCTCCTGTCACTGGTTATCACCCTTTACTGCAAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTGAGAGTGAAGTTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCCCCTCGCTAA
55 in SEQ ID NO: NT 5-DESIGN-NYESO 1-TCRba CD28 tmBBZ-Xho-NY-ESO 1
bmu2CD28tmBBcytCD3Zcyt-P2A-1g4TCRa95LYCD28tmBBcytCD3Zcyt-not. xdna nucleotide sequence
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACGGCTCTCCTAAGTTCTGGGTTCTCGTCGTCGTGGGAGGTGTGTTAGCATGTTACTCTCTCTTGGTTACTGTCGCTTTCATAATCTTTTGGGTCAAGAGAGGGCGTAAAAAGCTGCTCTACATCTTTAAGCAGCCTTTCATGCGTCCTGTTCAGACAACACAGGAAGAGGACGGATGCTCTTGCAGGTTCCCTGAGGAGGAGGAGGGTGGTTGCGAGCTCCGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGAGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCGGCTCCCCAAAATTTTGGGTGCTGGTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATTTTCTGGGTGAAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTGAGAGTGAAGTTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCCCCTCGCTAA
SEQ ID NO:56:NT6_NYESO1-TCRba28BBZ_Xho-NY-ESO1
bmu2 nucleotide sequence of CD28 tmcytbBytCD 3Zcyt-P2A-1g4TCRa95LYCD28 tmcytbBytCD 3Zcyt-not
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACGGCTCTCCTAAGTTCTGGGTTCTCGTCGTCGTGGGAGGTGTGTTAGCATGTTACTCTCTCTTGGTTACTGTCGCTTTCATAATCTTTTGGGTCCGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGTAAGAGAGGGCGTAAAAAGCTGCTCTACATCTTTAAGCAGCCTTTCATGCGTCCTGTTCAGACAACACAGGAAGAGGACGGATGCTCTTGCAGGTTCCCTGAGGAGGAGGAGGGTGGTTGCGAGCTCCGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGAGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCGGCTCCCCAAAATTTTGGGTGCTGGTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATTTTCTGGGTGAGGAGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCCCGCCGCCCCGGGCCCACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCCTATCGCTCCAAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTGAGAGTGAAGTTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCCCCTCGCTAA
57 in SEQ ID NO: NT21_ NT2-4h1_ NYESO1-TCRb28tmcytZa8tmBZh1_ Xho-NY-ESO1bmu2CD28tmcytCD3Zcyt-P2A-1g4TCRa95LYCD 8tmcytCD 3Zcyt-not. xBBna nucleotide sequence
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACGGCTCTCCTAAGTTCTGGGTTCTCGTCGTCGTGGGAGGTGTGTTAGCATGTTACTCTCTCTTGGTTACTGTCGCTTTCATAATCTTTTGGGTCCGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGTCGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGAGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCGGCTCCCCAAAATGTGATATCTACATCTGGGCGCCCTTGGCCGGGACTTGTGGGGTCCTTCTCCTGTCACTGGTTATCACCCTTTACTGCAAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTGAGAGTGAAGTTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCCCCTCGCTAA
58 in SEQ ID NO: NT22_ NT2-4h2_ NYESO1-TCRb28tmcytZa8tmBh2_ Xho-NY-ESO1bmu2CD28tmcytCD3Zcyt-P2A-1g4TCRa95LYCD8tmBBcyt-not
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACGGCTCTCCTAAGTTCTGGGTTCTCGTCGTCGTGGGAGGTGTGTTAGCATGTTACTCTCTCTTGGTTACTGTCGCTTTCATAATCTTTTGGGTCCGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGTCGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGAGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCGGCTCCCCAAAATGTGATATCTACATCTGGGCGCCCTTGGCCGGGACTTGTGGGGTCCTTCTCCTGTCACTGGTTATCACCCTTTACTGCAAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTGTAA
59 of SEQ ID NO: NT23_ NT2-5h1_ NYESO1-TCRb28tmcytZa28tmBZh1_ Xho-NY-ESO1bmu2CD28tmcytCD3Zcyt-P2A-1g4TCRa95LYCD28tmcytCD 3Zcyt-not. xBBna nucleotide sequence
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACGGCTCTCCTAAGTTCTGGGTTCTCGTCGTCGTGGGAGGTGTGTTAGCATGTTACTCTCTCTTGGTTACTGTCGCTTTCATAATCTTTTGGGTCCGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGTCGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGAGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCGGCTCCCCAAAATTTTGGGTGCTGGTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATTTTCTGGGTGAAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTGAGAGTGAAGTTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCCCCTCGCTAA
60 of SEQ ID NO: NT24_ NT2-4h3_ aNY-TCR-CAR b8tm28cytZa8tmBBcytZ _ Xho-NY-ESO1bmuCD8tmmu2CD28 cytuCD 3 Zcyt-linker
P2A-1g4TCRa95LYCD8tmBBcytZcyt-not
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACGGCTCTCCTAAGTGCGACATTTATATTTGGGCCCCTCTCGCTGGCACATGCGGCGTGTTGTTGCTCAGCCTCGTGATTACACTTTATTGTCGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGTCGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGAGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCGGCTCCCCAAAATGTGATATCTACATCTGGGCGCCCTTGGCCGGGACTTGTGGGGTCCTTCTCCTGTCACTGGTTATCACCCTTTACTGCAAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTGAGAGTGAAGTTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCCCCTCGCTAA
61: NT25_ aNY-TCR-CAR b8tm28 cytcty-a 8 tmBBcytyZ _ Xho-NY-ESO1bmu2CD8tmCD28 cytu 2CD3 Zcyt-linker
P2A-1g4TCRa95LYCD8 tmmuBBcyBcyBytZcyt-not
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACGGCTCTCCTAAGTGCGACATTTATATTTGGGCCCCTCTCGCTGGCACATGCGGCGTGTTGTTGCTCAGCCTCGTGATTACACTTTATTGTAGGAGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCCCGCCGCCCCGGGCCCACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCCTATCGCTCCCGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGTCGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGAGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCGGCTCCCCAAAATGTGATATCTACATCTGGGCGCCCTTGGCCGGGACTTGTGGGGTCCTTCTCCTGTCACTGGTTATCACCCTTTACTGCAAGAGAGGGCGTAAAAAGCTGCTCTACATCTTTAAGCAGCCTTTCATGCGTCCTGTTCAGACAACACAGGAAGAGGACGGATGCTCTTGCAGGTTCCCTGAGGAGGAGGAGGGTGGTTGCGAGCTCAAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTGAGAGTGAAGTTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCCCCTCGCTAA
62 of SEQ ID NO: NT26_ aNY-TCR-CARB8h28pectmcytZa8h28pectmcytZ _ Xho-NY-ESO1bmu2CD8hmu2CD28 pectmcyttmu 2Zcyt-P2A-1g nucleotide sequence of 4TCRa95LYCD8hCD28pectmcytZcyt-not
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACGGCTCTCCTAAGGCAAAACCGACGACCACCCCTGCCCCCAGGCCTCCTACTCCCGCCCCGACGATTGCCAGCCAACCGTTAAGTTTAAGACCGGAAGCATGTAGACCGGCAGCTGGTGGGGCTGTTCATACACGTGGCTTAGATTTTGCGCCTAGGAAGATCGAGGTAATGTACCCACCGCCCTATCTTGATAACGAAAAATCTAACGGTACAATAATTCACGTCAAGGGCAAGCATTTGTGCCCTTCCCCGTTGTTCCCGGGCCCAAGCAAACCGTTCTGGGTTCTCGTCGTCGTGGGAGGTGTGTTAGCATGTTACTCTCTCTTGGTTACTGTCGCTTTCATAATCTTTTGGGTCCGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGTCGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGAGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCGGCTCCCCAAAAGCTAAGCCCACCACGACGCCAGCGCCGCGACCACCAACACCGGCGCCCACCATCGCGTCGCAGCCCCTGTCCCTGCGCCCAGAGGCGTGCCGGCCAGCGGCGGGGGGCGCAGTGCACACGAGGGGGCTGGACTTCGCCCCTAGGAAAATTGAAGTTATGTATCCTCCTCCTTACCTAGACAATGAGAAGAGCAATGGAACCATTATCCATGTGAAAGGGAAACACCTTTGTCCAAGTCCCCTATTTCCCGGACCTTCTAAGCCCTTTTGGGTGCTGGTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATTTTCTGGGTGAGGAGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCCCGCCGCCCCGGGCCCACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCCTATCGCTCCAGAGTGAAGTTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCCCCTCGCTAA
SEQ ID NO:63:NT27_NT2b-4h1_aNY-TCR-CAR
Nucleotide sequence of bCD8h28pectmcytZa8tmBBcyt _ Xho-NY-ESO1bmu2CD8hmu2CD28pectmcytmu2Zcyt-P2A-1g4TCRa95LYCD8tmBBcyt-not
ATGAGCATCGGCCTCCTGTGCTGTGCAGCCTTGTCTCTCCTGTGGGCAGGTCCAGTGAATGCTGGTGTCACTCAGACCCCAAAATTCCAGGTCCTGAAGACAGGACAGAGCATGACACTGCAGTGTGCCCAGGATATGAACCATGAATACATGTCCTGGTATCGACAAGACCCAGGCATGGGGCTGAGGCTGATTCATTACTCAGTTGGTGCTGGTATCACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAGTTACGTCGGGAACACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACCGTACTGGAGGACCTGAAAAACGTGTTCCCACCCGAGGTCGCTGTGTTTGAGCCATCAGAAGCAGAGATCTCCCACACCCAAAAGGCCACACTGGTGTGCCTGGCCACAGGCTTCTACCCCGACCACGTGGAGCTGAGCTGGTGGGTGAATGGGAAGGAGGTGCACAGTGGGGTCAGCACAGACCCGCAGCCCCTCAAGGAGCAGCCCGCCCTCAATGACTCCAGATACGCTCTGAGCAGCCGCCTGAGGGTCTCGGCCACCTTCTGGCAGGACCCCCGCAACCACTTCCGCTGTCAAGTCCAGTTCTACGGGCTCTCGGAGAATGACGAGTGGACCCAGGATAGGGCCAAACCCGTCACCCAGATCGTCAGCGCCGAGGCCTGGGGTAGAGCAGACGGCTCTCCTAAGGCAAAACCGACGACCACCCCTGCCCCCAGGCCTCCTACTCCCGCCCCGACGATTGCCAGCCAACCGTTAAGTTTAAGACCGGAAGCATGTAGACCGGCAGCTGGTGGGGCTGTTCATACACGTGGCTTAGATTTTGCGCCTAGGAAGATCGAGGTAATGTACCCACCGCCCTATCTTGATAACGAAAAATCTAACGGTACAATAATTCACGTCAAGGGCAAGCATTTGTGCCCTTCCCCGTTGTTCCCGGGCCCAAGCAAACCGTTCTGGGTTCTCGTCGTCGTGGGAGGTGTGTTAGCATGTTACTCTCTCTTGGTTACTGTCGCTTTCATAATCTTTTGGGTCCGCTCAAAACGCTCTCGCTTGTTACATTCCGATTATATGAATATGACACCTAGGAGACCTGGCCCGACTAGGAAACACTATCAACCTTACGCACCTCCCAGAGATTTTGCTGCTTACAGGAGTCGGGTCAAATTTTCACGCTCCGCTGATGCTCCTGCCTATCAACAAGGGCAAAATCAATTGTACAATGAATTGAACTTGGGTAGAAGGGAAGAATATGACGTGCTCGATAAACGGAGGGGGAGAGATCCAGAAATGGGCGGTAAACCACGGCGCAAAAATCCACAAGAGGGATTGTATAACGAGCTCCAAAAGGACAAAATGGCAGAAGCTTATTCAGAAATAGGAATGAAGGGGGAAAGGAGACGAGGTAAAGGTCATGACGGATTGTATCAAGGATTGTCAACCGCTACTAAAGATACATATGATGCTTTGCATATGCAAGCTTTGCCTCCCAGAGCCAAGCGGTCTGGGTCTGGGGCCACCAACTTCAGCCTGCTGAAGCAGGCCGGCGACGTGGAGGAGAACCCCGGCCCCATGGAGACCCTCTTGGGCCTGCTTATCCTTTGGCTGCAGCTGCAATGGGTGAGCAGCAAACAGGAGGTGACGCAGATTCCTGCAGCTCTGAGTGTCCCAGAAGGAGAAAACTTGGTTCTCAACTGCAGTTTCACTGATAGCGCTATTTACAACCTCCAGTGGTTTAGGCAGGACCCTGGGAAAGGTCTCACATCTCTGTTGCTTATTCAGTCAAGTCAGAGAGAGCAAACAAGTGGAAGACTTAATGCCTCGCTGGATAAATCATCAGGACGTAGTACTTTATACATTGCAGCTTCTCAGCCTGGTGACTCAGCCACCTACCTCTGTGCTGTGAGGCCCCTGTACGGAGGAAGCTACATACCTACATTTGGAAGAGGAACCAGCCTTATTGTTCATCCGTATATCCAGAACCCTGACCCTGCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGAAAGTTCCGGCTCCCCAAAATGTGATATCTACATCTGGGCGCCCTTGGCCGGGACTTGTGGGGTCCTTCTCCTGTCACTGGTTATCACCCTTTACTGCAAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTGTAA
Compositions and kits
In one aspect, the present disclosure provides a composition comprising a plurality of genetically modified lymphocytes expressing a chimeric antigen receptor or chain thereof as described above for modulating the immune system of a subject.
Various lymphocytes can be used in the present invention. Examples of lymphocytes may include T cells, B cells, NK cells, macrophages, neutrophils, dendritic cells, mast cells, eosinophils, and basophils. In some embodiments, the lymphocytes are from CD34 hematopoietic stem cells, embryonic stem cells, or induced pluripotent stem cells. Lymphocytes may be autologous, allogeneic, syngeneic or xenogeneic. In some embodiments, the lymphocytes are autologous. In some embodiments, the lymphocyte is a human lymphocyte.
In some embodiments, the lymphocyte is a Peripheral Blood Lymphocyte (PBL). In some embodiments, the lymphocyte can be a Tumor Infiltrating Lymphocyte (TIL). In some embodiments, the lymphocytes can express a chimeric antigen receptor. In some embodiments, the lymphocytes may express a recombinant T cell receptor. The CAR or TCR can bind to a cancer antigen of interest.
Examples of such cancer and/or tumor antigens include, but are not limited to, for example, cyclin-dependent kinase-4, beta-catenin, Caspase-8, MAGE-1, MAGE-3, tyrosinase, surface Ig idiotypes, Her-2/neu, MUC-1, HPV E6, HPV E7, CD5, idiotypes, CAMPATH-1, CD20, CEA, mucin-1, Lewisx、CA-125、EGFR、p185HER2IL-2R, FAP, tenascin, metalloprotease, phCG, gp100 or Pmlll 7, HER2/neu, CEA, gp100, MART1, TRP-2, melanin-A, NY-ESO-1, MN (gp250), idiotype, MAGE-1, MAGE-3, tyrosinase, telomerase, MUC-1 antigen and germ cell derived antigens, blood group antigens, e.g., Lea, Leb, LeX, LeY, H-2, B-1, B-2 antigens.
In certain embodiments, T cells expressing the same CAR can bind to more than one cancer and/or tumor antigen; for example, binding of one CAR of a T cell to a MAGE antigen can be combined with binding of another CAR antigen of a T cell (such as melanin a, tyrosinase, or gp 100). For example, CD20 is a pan B antigen present on both malignant and non-malignant B cell surfaces that has been shown to be a very effective target for immunotherapeutic antibodies to treat non-hodgkin's lymphoma. In this respect, pan T cell antigens (such as CD2, CD3, CD5, CD6 and CD7) also comprise tumor-associated antigens within the meaning of the present invention. Other exemplary tumor associated antigens include, but are not limited to, MAGE-1, MAGE-3, MUC-1, HPV 16, HPV E6& E7, TAG-72, CEA, L6-antigen, CD19, CD22, CD37, CD52, HLA-DR, EGF receptor, and HER2 receptor. In many cases, immunoreactive antibodies (and/or immunoreactive antigen-binding fragments) against each of these antigens have been reported in the literature.
In embodiments, both strands of the chimeric antigen receptor are encoded by one nucleic acid transgene. The two strands may be linked by a self-cleaving peptide sequence. Alternatively, the two sequences encoding the two strands are linked by a nucleic acid sequence comprising an IRES such that the two strands can be translated separately. Expression of the transgene may be regulated by a constitutively active promoter or by an inducible promoter. In some embodiments, expression of the transgene may be induced by the activation state of lymphocytes. In other cases, transgenes can be introduced into lymphocytes by gamma retroviruses or lentiviruses with integration capability, DNA transposition, and the like.
The genetically modified lymphocytes described above can be introduced into a pharmaceutical composition suitable for administration. The pharmaceutical composition typically comprises substantially isolated/pure lymphocytes and a pharmaceutically acceptable carrier in a form suitable for administration to a subject. Pharmaceutically acceptable carriers can be determined, in part, by the particular composition being administered and the particular method used to administer the composition. Pharmaceutical compositions are typically formulated to fully comply with the Good Manufacturing Practice (GMP) regulations of the U.S. food and drug administration.
The term "pharmaceutically acceptable" in reference to compositions, carriers, diluents and agents is used interchangeably and includes substances that are capable of being administered to a subject or that do not produce undesirable physiological effects after administration to a subject to the extent that they would prevent administration of the composition. For example, "pharmaceutically acceptable excipients" include excipients that are generally safe, non-toxic and desirable for use in the preparation of pharmaceutical compositions, and include acceptable excipients for veterinary as well as human use.
Examples of such carriers or diluents include, but are not limited to, water, saline, ringer's solution, dextrose solution, and 5% human serum albumin. The use of such media and compounds for pharmaceutically active substances is well known in the art. Unless any conventional media or compound is incompatible with the disclosed compositions, it is contemplated that it can be used in the compositions. In some embodiments, a second therapeutic agent (e.g., anti-cancer or anti-tumor) may also be added to the pharmaceutical composition.
Pharmaceutical compositions suitable for injectable use include sterile aqueous solutions (where water soluble) or dispersions and sterile powders for the extemporaneous preparation of sterile injectable solutions or dispersions. To pairFor intravenous administration, suitable carriers include physiological saline, bacteriostatic water, Cremophor ELTM(BASF, Parsippany, n.j.) or Phosphate Buffered Saline (PBS). In all cases, the composition must be sterile and should be fluid to the extent that easy injection is possible. It must be stable under the conditions of manufacture and storage and must be preserved against microbial (e.g., bacterial and fungal) contamination. The carrier can be a solvent or dispersion medium containing, for example, water, ethanol, polyol (for example, glycerol, propylene glycol, and liquid polyethylene glycol, and the like), and suitable mixtures thereof. Proper fluidity can be maintained, for example, by the use of a coating such as lecithin, by the maintenance of the required particle size in the case of dispersions and by the use of surfactants. In some embodiments, the composition comprises genetically modified lymphocytes as described above and optionally a lyophilization protectant (e.g., glycerol, DMSO, PEG).
The compositions or pharmaceutical compositions described herein may be provided in a kit. In one embodiment, the kit comprises (a) a container containing the composition and optionally (b) informational material. The informational material may be descriptive, marketing, or other material related to the use of the methods and/or agents described herein for therapeutic benefit. For example, the kit may include instructions for manufacture, for the treatment regimen and the administration period to be used. In one embodiment, the kit may further comprise an additional therapeutic agent (e.g., a checkpoint modulator). The kit may comprise one or more containers, each containing a different agent. For example, a kit comprises a first container containing the composition and a second container containing an additional therapeutic agent.
The container may contain a unit dose of the pharmaceutical composition. In addition to the composition, the kit may contain other ingredients such as solvents or buffers, adjuvants, stabilizers or preservatives. The kit optionally comprises a device suitable for administering the composition, e.g., a syringe or other suitable delivery device. The device may be pre-loaded with one or both medicaments, or may be empty but suitable for loading.
Process for preparing a composition
In general, the practice of the present invention employs, unless otherwise indicated, conventional techniques of chemistry, molecular biology, recombinant DNA techniques, PCR techniques, immunology (e.g., antibody techniques), expression systems (e.g., cell-free expression, phage display, ribosome display, and PROFUSION), and any necessary cell culture within the skill of the art and as set forth in the literature. Although certain aspects of the invention relate to compositions and uses of recombinant RNA retroviruses (e.g., lentiviruses HIV-2, SIV, etc.), molecular cloning can be performed using proviral DNA cloning, thereby allowing standard cloning techniques to be used. Site-directed mutagenesis in vitro by synthesis of oligodeoxynucleotides may be performed according to methods well known in the art. The Gene fusions of the invention (particularly for use in the synthesis of fusion proteins, such as CARs) can be prepared by methods well known in the art, for example, the Gene SOE (by overlap extension splicing) method (Horton et al, 1989Gene 77: 61-68; U.S. Pat. No. 5,023,171) which typically relies on the use of fusion primers (which are optionally mutagenic) during PCR amplification. Enzymatic amplification of DNA fragments can be carried out by PCR techniques using a DNA thermal cycler according to the manufacturer's instructions.
Verification of the nucleotide sequence can be performed by sequencing. Whether a homologous recombination event has occurred between two homologous polypeptides contained in a single vector of the invention can be verified by any method known in the art, including, but not limited to, Northern blot and/or RT-PCR methods (e.g., if assessed directly within an isolated retroviral genome), Southern blot and/or PCR methods (e.g., assessing host cell genomic DNA containing an integrated retroviral vector), and SDS-PAGE followed by Western blot and/or immunoprecipitation followed by SDS-PAGE and detection of labeled polypeptides (e.g., if the homologous polypeptides are of a discernible size and/or contain distinguishable domains, features, and/or epitopes)
Some embodiments relate to the creation of retroviral vectors encoding two or more identical or highly homologous molecules with degenerate codons. Molecular Therapy-Methods & Clinical Development (2014) article numbers have been previously reported in, for example, U.S. patent numbers 9,206,440 and Im EJ et al, combination-deletion between homology cassettes in retroviruses is supplied by a strain of production code subscription: 14022 describes a method of using silent mutation to create a retroviral vector that encodes two or more identical or highly homologous molecules with degenerate codons to reduce possible DNA recombination events. The references are incorporated by reference.
In another aspect, the present disclosure provides a method of making the composition described above. The method comprises the following steps: (a) providing a plurality of lymphocytes; (b) introducing a nucleic acid molecule encoding a first polypeptide chain and a second polypeptide chain into a plurality of lymphocytes to obtain a plurality of genetically engineered lymphocytes; and (c) amplifying the plurality of genetically engineered lymphocytes in cell culture medium.
In some embodiments, the method may comprise: (a) providing a plurality of lymphocytes; (b) introducing a first nucleic acid and a second nucleic acid encoding a first polypeptide chain and a second polypeptide chain, respectively, into a plurality of lymphocytes, thereby obtaining a plurality of genetically engineered lymphocytes; and (c) amplifying the plurality of genetically engineered lymphocytes in cell culture medium. In some embodiments, the method can further comprise expanding the plurality of first lymphocytes in the cell culture medium after the step of introducing the first nucleic acid, or expanding the plurality of second lymphocytes in the cell culture medium after the step of introducing the second nucleic acid.
The method of obtaining the composition of the tumor-specific genetically modified subset of lymphocytes described above can be performed in vitro or ex vivo. A more specific form of the process may be as disclosed in PCT/EP2018/080343, the contents of which are incorporated herein by reference in their entirety.
The term "culturing" or "expanding" refers to maintaining or culturing cells under conditions in which the cells can proliferate and avoid senescence. For example, the cells can be cultured in a medium that optionally includes one or more growth factors (i.e., a mixture of growth factors). In some embodiments, the cell culture medium is a defined cell culture medium. The cell culture medium may comprise a neoantigenic peptide. Stable cell lines can be established to allow for continued propagation of the cells.
Lymphocytes
A source of lymphocytes from the subject is obtained prior to expansion and genetic modification of the lymphocytes. Lymphocytes can be from several sources, including peripheral blood mononuclear cells, bone marrow, lymph node tissue, cord blood, thymus tissue, tissue from the site of infection, ascites, pleural effusion, spleen tissue, and tumors. Any number of lymphocyte cell lines available in the art can be used, as described herein. Various techniques known to those skilled in the art (e.g., Ficoll) can be usedTMIsolated), lymphocytes are obtained from a unit of blood collected from the subject. Circulating blood cells of an individual may be obtained by apheresis. Apheresis products typically contain lymphocytes, including T lymphocytes, monocytes, granulocytes, B lymphocytes, other nucleated leukocytes, erythrocytes, and platelets. Cells collected by apheresis may be washed to remove plasma fractions and placed in an appropriate buffer or culture medium for subsequent processing steps. Cells can be washed with PBS. Alternatively, the wash solution may contain no calcium and no magnesium, or may contain many, if not all, divalent cations. One of ordinary skill in the art will appreciate that the washing step can be accomplished by methods well known to those skilled in the art, such as using a semi-automatic continuous flow centrifuge (e.g., Cobe 2991 Cell processor, the Baxter CytoMate or e1Haemonetics Cell Saver 5) according to the manufacturer's instructions. After washing, the cells can be resuspended in various biocompatible buffers, such as, for example, Ca2+ free, Mg2+ free PBS, PlasmaLyte a, or other salt solutions with or without buffers. Alternatively, the apheresis sample may be freed of unwanted components and the cells resuspended directly in culture.
Lymphocytes are isolated from peripheral blood by lysing erythrocytes and depleting monocytes, for example, by PERCOLL gradient centrifugation or countercurrent centrifugation elutriation, as described herein. If desired, a particular subpopulation of lymphocytes, such as T lymphocytes (i.e., Cd3+, Cd28+, Cd4+, Cd8+, Cd45RA +, or Cd45RO + T lymphocytes), may be further isolated by positive or negative selection techniques. For example, the desired T lymphocytes can be positively selected by isolating the T lymphocytes using conjugated anti-CD 3/anti-CD 28 beads (i.e., 3x28), such as DYNABEADS M-450CD3/CD 28T, incubated for a sufficient period of time (i.e., 30 minutes to 24 hours). For the isolation of T lymphocytes from leukemia patients, the use of longer incubation times (e.g., 24 hours) can improve cell performance. In any case where T lymphocytes are rare compared to other cell types, such as in any case where TIL is isolated from tumor tissue or immunocompromised individuals, longer incubation times can be used to isolate T lymphocytes. Those skilled in the art will appreciate that multiple rounds of selection may also be used. It may be desirable to perform a selection procedure and use "unselected" cells during activation and expansion. A new round of selection can also be performed on "unselected" cells.
The enriched lymphocyte (e.g., T lymphocyte) population can be negatively selected by combining antibodies directed against unique surface markers of the negatively selected cells. One approach is to sort and/or select cells by negative magnetic immunoadhesion or flow cytometry by using a mixture of monoclonal antibodies directed against cell surface markers present in the negatively selected cells. For example, to enrich for CD4+ cells by negative selection, monoclonal antibodies typically include antibodies against CD14, CD20, CD11b, CD16, HLA-DR, and CD 8. Alternatively, regulatory T lymphocytes are depleted by anti-C25 conjugate beads or other similar selection methods.
Lymphocytes for stimulation may also be frozen after the washing step. Without wishing to be bound by theory, the freezing and subsequent thawing steps provide a more uniform product by eliminating granulocytes and to some extent monocytes in the cell population. After a washing step to remove plasma and platelets, the cells may be suspended in a freezing solution. Although many solution and freezing parameters are known in the art and will be useful in this context, one approach involves the use of PBS containing 20% DMSO and 8% human serum albumin, or a medium containing 10% dextran 40 and 5% dextrose human albumin and 7.5% DMSO or 31.25% plasma a, 31.25% dextrose 5%, 0.45% NaCl, 10% dextran 40 and 5% dextrose, 20% human albumin serum and 7.5% DMSO or other suitable cell freezing medium containing, for example, Hespan and PlasmaLyte a. The cells can then be frozen at-80 ℃ at a rate of 1 ℃ per minute and stored in the gas phase of a liquid nitrogen storage tank. Other methods of controlled freezing may be used, as well as immediate uncontrolled freezing at-20 ℃ or in liquid nitrogen.
Frozen cells can be thawed and washed as described herein and allowed to stand at room temperature for one hour prior to activation using the methods of the invention. As described herein, lymphocytes can be expanded, frozen, and used at a later time. As described herein, a sample may be collected from a patient shortly after diagnosis of a particular disease as described herein, but prior to any treatment. Cells can be isolated from a subject's blood sample or apheresis prior to any number of relevant treatment modalities, including, but not limited to, the use of agents (e.g., natalizumab, efuzumab), antiviral agents, chemotherapy, radiation, immunosuppressive agents (e.g., cyclosporine, azathioprine, methotrexate, mycophenolate, and FK506), antibodies or other immunoablative agents (e.g., camp, anti-CD 3 antibodies, cytoxane, fludarabine, cyclosporine, FK506, rapamycin, mycophenolic acid, steroids, FR901228, and radiation). These drugs inhibit calcium-dependent calcineurin (e.g., cyclosporin and FK506) or inhibit p70S6 kinase, which is important for growth factor (rapamycin) induced signaling (Liu et al, Cell 66:807-815, 1991; Henderson et al, Immun 73:316-321,199, Bierer et al, curr. Opin. Immun.,5:763-773, 1993). Cells may be isolated from a patient and frozen for subsequent use (e.g., prior to, concurrently with, or subsequent to) bone marrow or stem cell transplantation, T lymphocyte ablation therapy with chemotherapeutic agents (e.g., fludarabine), radiation external to radiotherapy (XRT), cyclophosphamide, or antibodies (e.g., OKT3 or CAMPATH). As described herein, cells can be isolated prior and can be frozen for treatment following B lymphocyte ablation therapy (such as agents that react with CD20, e.g., rituximab).
Prior to or after genetic modification of lymphocytes to express the desired transgene, one can generally use a transgene such as, for example, those described in U.S. patent 6,352,694; 6,534,055, respectively; 6,905,680, respectively; 6,692,964, respectively; 5,858,358, respectively; 6,887,466, respectively; 6,905,681, respectively; 7,144,575, respectively; 7,067,318, respectively; 7,172,869, respectively; 7,232,566, respectively; 7,175,843, respectively; 5,883,223, respectively; 6,905,874, respectively; 6,797,514, respectively; 6,867,041, respectively; and those described in U.S. patent application 20060121005 to activate and expand lymphocytes.
Carrier
Various methods can be used to introduce transgenes into lymphoid cells. These methods include, but are not limited to, transduction of cells using integrating gamma retrovirus or lentivirus and DNA transposition.
A variety of vectors can be used for expression of the transgene. The ability of certain viruses to infect or enter cells via receptor-mediated endocytosis, integrate into the host cell genome and stably and efficiently express viral genes, making them attractive candidates for transferring foreign nucleic acids into cells. Thus, in certain embodiments, the viral vector is used to introduce into a host cell for expression a nucleotide sequence encoding one or more transgenes or fragments thereof. The viral vector may comprise a nucleotide sequence encoding one or more transgenes or fragments thereof operably linked to one or more control sequences (e.g., a promoter). Alternatively, the viral vector may not contain control sequences, but rather rely on control sequences within the host cell to drive expression of the transgene or fragment thereof. Non-limiting examples of viral vectors that can be used to deliver the nucleic acid include adenoviral vectors, AAV vectors, and retroviral vectors.
For example, adeno-associated virus (AAV) can be used to introduce nucleotide sequences encoding one or more transgenes or fragments thereof into a host cell for expression. AAV systems have been described previously and are generally well known in the art (Kelleher and Vos, Biotechniques,17(6): 1110-. Details of the production and use of rAAV vectors are described, for example, in U.S. patent nos. 5,139,941 and 4,797,368, which are all incorporated by reference herein in their entirety for all purposes.
In some embodiments, retroviral expression vectors can be used to introduce nucleotide sequences encoding one or more transgenes or fragments thereof into a host cell for expression. These systems have been described previously and are generally well known In the art (Nicolas and Rubinstein, In, Rodriguez and Denhardt, Stoneham: Butterworth, pp.494-513,1988; Temin, In: Gene Transfer, Kucherlapati (ed.), New York: Plenum Press, pp.149-188,1986). Examples of vectors for eukaryotic expression in mammalian cells include AD5, pSVL, pCMV, pRc/RSV, pcDNA3, pBPV, etc., and vectors derived from viral systems (e.g., vaccinia virus, adeno-associated virus, herpes virus, retrovirus, etc.) using promoters (e.g., CMV, SV40, EF-1, UbC, RSV, ADV, BPV, and β -actin).
Combinations of retroviruses and suitable packaging cell lines may also find use where the capsid proteins will function to infect target cells. Typically, the cells and virus are incubated in the medium for at least about 24 hours. Then, prior to analysis, in some applications, the cells are allowed to grow in culture for a short time (e.g., 24-73 hours) or at least two weeks, and the cells may be allowed to grow for five weeks or more. Commonly used retroviral vectors are "defective", i.e., incapable of producing the viral proteins required for productive infection. Replication of the vector requires growth in a packaging cell line. The host cell specificity of retroviruses is determined by the envelope protein env (pl 20). The envelope protein is provided by a packaging cell line. Envelope proteins are of at least three types, ecotropic, amphotropic and heterophilic. Retroviruses (e.g., MMLV) packaged with ecotropic envelope proteins are capable of infecting most mouse and rat cell types. The heterotrophic packaging cell line includes BOSC 23. Retroviruses carrying the amphipathic envelope protein (e.g., 4070A) are capable of infecting most mammalian cell types, including humans, dogs and mice. The amphipathic packaging cell line includes PA12 and PA 317. Retroviruses (e.g., AKR env) packaged with xenotropic envelope proteins are capable of infecting most mammalian cell types, except mouse cells. The vector may contain genes that must be subsequently removed, for example using a recombinase system (such as Cre/Lox), or the cells expressing it may be disrupted, for example by containing genes that allow selective virulence (such as herpes virus TK, BCL-xs, etc.). A suitable inducible promoter is activated in the desired target cell type (i.e., the transfected cell or progeny thereof).
Non-limiting examples of useful vectors include the retroviral vector SFG. MCS and helper plasmids RD114, Peg-Pam3(Arber et al, J Clin Invest 2015Jan 2; 125(1): 157-. In some embodiments, a sleeping beauty swivel system (Deniger et al, 2016Mol ther. Jun; 24(6):1078 and 1089) may be used. In some embodiments, the transgene is introduced into the Cell by passing it through a small opening, deforming the Cell, disrupting the Cell membrane and inserting material into the Cell, e.g., electroporation (Xiaojun et al, 2017Protein Cell,8(7): 514) or cellsA method. Such electroporation methods of RNA encoding transgenes can transiently express such transgenes in cells, thereby limiting toxicity and other adverse effects of engineered cells (Barrett et al, 2011Hum Gene Ther. Dec; 22(12): 1575-1586).
Physical methods for introducing polynucleotides into host cells include calcium phosphate precipitation, lipofection, particle bombardment, microinjection, electroporation, and the like. Methods for producing cells comprising exogenous vectors and/or nucleic acids are well known in the art. See, e.g., Sambrook et al (2001, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, New York).
Chemical methods for introducing polynucleotides into host cells include colloidally dispersed systems such as macromolecular complexes, nanocapsules, microspheres, beads and lipid-based systems including oil-in-water emulsions, micelles, mixed micelles and liposomes. Exemplary colloidal systems for use as vehicles for in vitro and in vivo release are liposomes (e.g., artificial membrane vesicles).
In the case where a non-viral delivery system is used, an exemplary delivery vehicle is a liposome. Relates to the use of lipid formulations for introducing nucleic acids into host cells (in vitro, ex vivo or in vivo). In another aspect, the nucleic acid can be bound to a lipid. The nucleic acid associated with the lipid may be encapsulated in the aqueous interior of the liposome, dispersed within the lipid bilayer of the liposome, associated with the liposome via a binding molecule that is associated with both the liposome and the oligonucleotide, encapsulated in the liposome, formed into a complex with the liposome, dispersed in a solution containing the lipid, mixed with the lipid, combined with the lipid, included as a suspension in the lipid, the contents or in a complex with micelles, or otherwise associated with the lipid. The composition associated with the lipid, lipid/DNA or lipid/expression vector is not limited to any particular structure in solution. For example, it may exist in a double-layer structure in the form of micelles, or may be in a "collapsed" structure. It may also simply be dispersed in the solution, possibly forming aggregates that are not uniform in size or shape. Lipids are fatty substances, which may be natural or synthetic lipids. For example, lipids include fat droplets that naturally occur in the cytoplasm, and a class of compounds containing long-chain aliphatic hydrocarbons and their derivatives, such as fatty acids, alcohols, amines, amino alcohols, and aldehydes.
Lipids suitable for use can be obtained from commercial sources. For example, dimyristylphosphatidylcholine ("DMPC") is available from Sigma, st.louis, MO; hexacosanyl phosphate ("DCP") is available from K & K Laboratories (Plainview, NY); cholesterol ("Choi") is available from Calbiochem-Behring; dimyristylphosphatidylglycerol ("DMPG") and other Lipids are available from Avanti Polar Lipids, Inc. Lipid stocks in chloroform or chloroform/methanol may be stored at about-20 ℃. Chloroform was used as the only solvent because it evaporates more readily than methanol. "liposomes" is a generic term that encompasses a variety of unique and multilamellar lipid carriers formed by the creation of bilayer or closed lipid aggregates. Liposomes are characterized by having a vesicular structure with a phospholipid bilayer membrane and an internal aqueous medium. Multilamellar liposomes have multiple lipid layers separated by an aqueous medium. Phospholipids form spontaneously when suspended in excess aqueous solution. The lipid component undergoes self-rearrangement before forming a closed structure and traps dissolved water and solutes between lipid bilayers (Ghosh et al, 1991Glycobiology 5: 505-10). However, compositions having a structure different from the normal vesicle structure in solution are also included. For example, lipids may have a micellar structure, or may simply exist as heterogeneous aggregates of lipid molecules. Also relates to liposome (Lipofectamine) -nucleic acid complexes.
Regardless of the method used to introduce the exogenous nucleic acid into the host cell, the presence of the recombinant DNA sequence in the host cell can be confirmed by a series of assays. Such assays include, for example, "molecular biology" assays well known to those skilled in the art, such as Southern and Northern blots, RT-PCR and PCR; biochemical assays, such as detecting the presence or absence of a particular peptide, for example, by immunological methods (ELISA and Western blot) or by assays described herein to identify agents within the scope of the invention.
Method of treatment
The agents (e.g., vectors and cells) described above can be used in immunotherapy for treating various diseases. Accordingly, the present disclosure also provides a method of treating a condition (such as an infection, cancer, or tumor). The method comprises administering to a subject in need thereof a therapeutically effective amount of a composition or pharmaceutical composition as described above.
As used herein, the terms "subject" and "patient" are used interchangeably regardless of whether the subject is receiving or is currently receiving any form of treatment. As used herein, the terms "subject" and "subjects" can refer to any vertebrate, including but not limited to mammals (e.g., cows, pigs, camels, llamas, horses, goats, rabbits, sheep, hamsters, guinea pigs, cats, dogs, rats and mice, non-human primates (e.g., monkeys, such as cynomolgus monkeys, chimpanzees, etc.), and humans). The subject may be human or non-human. In a more exemplary aspect, the mammal is a human. In some embodiments, the subject is a human. In some embodiments, the subject has cancer. In some embodiments, the subject is immunocompromised.
Cancer treatment
In some embodiments, the agents (e.g., vectors and cells) described above can be used in immunotherapy for treating cancer or tumors. As used herein, "cancer," "tumor," and "malignancy" all equally relate to the proliferation of a tissue or organ. If the tissue is part of the lymphatic or immune systems, the malignant cells may comprise non-solid tumors of circulating cells. Malignant tumors of other tissues or organs may produce solid tumors. The methods of the invention can be used to treat lymphocytes, circulating immune cells, and solid tumors.
Cancers that may be treated include non-vascularized or substantially non-vascularized tumors as well as vascularized tumors. The cancer may comprise a non-solid tumor (such as a hematological tumor, e.g., leukemia and lymphoma) or may comprise a solid tumor. The types of cancers that can be treated using the compositions of the present invention include, but are not limited to, carcinomas, blastomas and sarcomas, as well as certain leukemias or malignant lymphoid tumors, benign and malignant tumors, e.g., sarcomas, carcinomas, and melanomas. Also included are adult tumors/cancers and pediatric tumors/cancers.
The hematologic cancer is a hematologic or bone marrow cancer. Examples of hematologic (or hematological) cancers include leukemias, including acute leukemias (e.g., acute lymphocytic leukemia, acute myelocytic leukemia, acute myelogenous leukemia, promyelocytic, myelomonocytic, monocytic, and erythroleukemia), chronic leukemias (e.g., chronic myelocytic (myelocytic) leukemia, chronic myelogenous leukemia, and chronic lymphocytic leukemia), polycythemia vera, lymphoma, hodgkin's disease, non-hodgkin's lymphoma (indolent and high grade forms), multiple myeloma, fahrenheit macroglobulinemia, heavy chain disease, myelodysplastic syndrome, hairy cell leukemia, and myelodysplasia.
Solid tumors are abnormal tissue masses that generally do not contain cysts or fluid areas. Solid tumors can be benign or malignant. Different types of solid tumors are named for the cell types that form the solid tumor (e.g., sarcomas, carcinomas, and lymphomas). Examples of solid tumors (e.g., sarcomas and carcinomas) include fibrosarcoma, myxosarcoma, liposarcoma, chondrosarcoma, osteosarcoma, and other sarcomas, synovial membrane, mesothelioma, ewing's tumor, leiomyosarcoma, rhabdomyosarcoma, colon carcinoma, lymphoid malignancies, pancreatic cancer, breast cancer, lung cancer, ovarian cancer, prostate cancer, hepatocellular carcinoma, squamous cell carcinoma, basal cell carcinoma, adenocarcinoma, sweat gland carcinoma, medullary thyroid carcinoma, papillary thyroid carcinoma, sebaceous gland carcinoma of pheochromocytoma, papillary carcinoma, papillary adenocarcinoma, medullary carcinoma, bronchial carcinoma, renal cell carcinoma, liver cancer, bile duct carcinoma, choriocarcinoma, wilms' tumor, cervical cancer, testicular tumor, seminoma, bladder cancer, melanoma, and CNS tumors (e.g., gliomas (such as brain stem glioma and mixed gliomas), glioblastoma (also known as astrocytoma), CNS lymphoma, germ cell tumor, medulloblastoma, schwannoma, craniopharyngioma, ependymoma, pinealoma, hemangioblastoma, acoustic neuroma, oligodendroglioma, meningioma, neuroblastoma, retinoblastoma, and brain metastases.
The above agents can also be used as immunotherapeutic agents for the treatment of infectious diseases; for example, in a procedure using a CAR that recognizes an infectious disease antigen. Thus, polypeptides described herein can be prepared that bind to various forms of infectious disease antigens, thereby inducing an immune response to the infectious disease antigens upon binding. The CARs described herein can be designed to bind to infectious disease antigens including, but not limited to, bacterial antigens, viral antigens, fungal antigens, parasitic antigens, and microbial toxins. Exemplary forms of each class of antigen are considered in more detail below.
Bacteria
Examples of bacteria (in particular, epitopes thereof) to which a polypeptide or CAR of the invention can bind include, but are not limited to: pseudomonas aeruginosa (Pseudomonas aeruginosa), Pseudomonas fluorescens (Pseudomonas fluorescens), Pseudomonas putida (Pseudomonas acidovorans), Pseudomonas putida (Pseudomonas putida), Stenotrophomonas maltophilia), Burkholderia cepacia (Burkholderia cepacia), Aeromonas hydrophila (Aeromonas hydrophylla), Escherichia coli (Escherichia coli), Citrobacter freundii (Citrobacter freundii), Salmonella Typhimurium (Salmonella enterica Typhimurium), Salmonella Typhi (Salmonella Typhimurium), Salmonella Paratyphi (Salmonella choleraesuis), Salmonella Typhi (Salmonella choleraesula), Salmonella Typhimurium (Salmonella enterica), Salmonella Typhi (Salmonella choleraesula), Salmonella Typhimurium (Escherichia coli), Salmonella Typhimurium (Salmonella choleraesuis), Salmonella Typhimurium (Escherichia coli), Salmonella Typhimurium (Escherichia coli), Salmonella choleraesuis (Escherichia coli), Salmonella choleraesuis, Salmonella Enterobacter coli (Escherichia coli), Salmonella choleraesuis (Escherichia coli), Salmonella Enterobacter coli (Escherichia coli), Salmonella choleraesuis, Salmonella Typhimurium (Escherichia coli), Salmonella Typhimurium) Serratia marcescens (Serratia marcescens), Francisella tularensis (Francisella tularensis), Morganella morganii (Morganella morganii), Proteus mirabilis (Proteus mirabilis), Proteus vulgaris (Proteus vulgaris), Alkallidis alcaligenes (Providecia alcalifaciens), Providella rapae (Providecia rettgeri), Providencia stuartii (Providecia stuartii), Acinetobacter calcoaceticus (Acinetobacter calcoaceticus), Acinetobacter haemolyticus (Acinetobacter haementosus), Yersinia enterocolitica (Yersinia entomolytica), Yersinia pestis (Yersinia pestis), Yersinia pseudonarcoticina (Yersinia verruckeri), Yersinia verruckeri (Yersinia), Yersinia intermediate Yersinia, Yersinia parahaemophila (Bonderella), Hawthorn), Haematitum (Bordetectinospora, Hawthorn), Haemarrhoea, Haemarrhinula paradoxeria (Bovinifera), Hawthorn), Haemarrhinula (Bordetectinospora), Hawthia, Havinifera (Bordetectinospora, Havinifera), Havinifera (Bordetectina), Havinifera, Bordetecticola (Bordetectina), Bordetectina, haemophilus parahaemolyticus (Haemophilus parahaemolyticus), Haemophilus ducreyi (Haemophilus ducreyi), Pasteurella multocida (Pasteurella multocida), Pasteurella haemolyticus (Pasteurella haemolytica), Bacillus catarrhalis (Branhamella catarrhalis), Helicobacter pylori (Helicobacter pylori), Campylobacter (Campylobacter coli), Campylobacter jejuni (Campylobacter jejuni), Campylobacter coli (Campylobacter coli), Borrelia burgdorferi (Borrelia burgdorferi), Vibrio cholerae (Vibrio cholerae), Vibrio parahaemolyticus (Vibrio parahaemolyticus), Legionella pneumonia (Legionella parahaemophilus), Neisseria monocytogenes (Listeria meningitidis), Neisseria meningitidis (Salmonella parahaemolytica), Neisseria meningitidis (Salmonella parahaemolyticus), Neisseria meningitidis (Salmonella parahaemolyticus), Neisseria meningitidis (Salmonella parahaemolyticus), Neisseria meningitidis (Neisseria meningitidis), Neisseria meningitidis (Neisseria meningitidis), Neisseria, Bacteroides thetaiotaomicron (Bacteroides thetaiotaomicron), Bacteroides monoformans (Bacteroides uniformis), Bacteroides exserohilus (Bacteroides eggertii), Bacteroides endophyticus (Bacteroides stramenomyces), Clostridium difficile (Clostridium difficile), Mycobacterium tuberculosis (Mycobacterium tuberculosis), Mycobacterium avium (Mycobacterium avium), Mycobacterium intracellulare (Mycobacterium intracellulare), Mycobacterium leprae (Mycobacterium leprae), Corynebacterium diphtheriae (Corynebacterium diphtheriae), Mycobacterium ulcerobacter ulcerosus (Corynebacterium ulcerococcus ulorans), Streptococcus pneumoniae (Staphylococcus pneumoniae), Streptococcus agalactiae (Staphylococcus aureus), Streptococcus pyogenes (Staphylococcus aureus), Staphylococcus aureus (Staphylococcus epidermidis), Staphylococcus aureus (Staphylococcus aureus) Human Staphylococcus (Staphylococcus hominis) and Staphylococcus sacchari (Staphylococcus saccharolyticus). In a particular embodiment, the construct of the invention comprises a binding molecule that binds to staphylococcal protein a.
Virus
Examples of viruses (or epitopes thereof) to which a polypeptide or CAR of the invention can bind include, but are not limited to: polyoma virus JC (JCV), human immunodeficiency virus type I (HIV I), Hepatitis B Virus (HBV), Hepatitis C Virus (HCV), Cytomegalovirus (CMV), Epstein Barr Virus (EBV), influenza virus hemagglutinin (Genbank accession No. J02132; Air,1981, Proc. Natl. Acad. Sci. USA 78:7639-, Hepatitis B virus surface antigen (Itoh et al, 1986, Nature 308: 19; Neurath et al, 1986, Vaccine 4:34), diphtheria toxin (Audibert et al, 1981, Nature 289:543), Streptococcus 24M epitope (Beachey,1985, adv. Exp. Med. biol.185:193), gonococcal pilin (Rothbard and Schoolnik,1985, adv. Exp. Med. biol.185:247), pseudorabies virus g50(gpD), pseudorabies virus II (gpB), pseudorabies virus gIII (gpC), pseudorabies virus glycoprotein H, pseudorabies virus glycoprotein E, transmissible gastroenteritis glycoprotein 195, transmissible gastroenteritis matrix protein, porcine rotavirus glycoprotein 38, porcine parvovirus capsid protein, porcine dysentery spirochete protective antigen, bovine viral diarrhea glycoprotein 55, newcastle disease virus neuraminidase, porcine African neuraminidase, foot-and mouth disease virus, swine influenza virus hemagglutinin, swine influenza hemagglutinin, swine fever virus influenza virus hemagglutinin, swine fever virus infection, swine fever, swine, Mycoplasma hyopneumoniae, infectious bovine rhinotracheitis virus (e.g., infectious bovine rhinotracheitis virus glycoprotein E or glycoprotein G) or infectious laryngotracheitis virus (e.g., infectious laryngotracheitis virus glycoprotein G or glycoprotein I), glycoproteins of the Rake virus (Gonzales Scarano et al, 1982, Virology 120:42), neonatal calf diarrhea virus (Matsuno and Inouye,1983, Infection and Immunity 39:155), Venezuelan equine encephalitis virus (Mathews and Roehrigig, 1982, J.Immunol.129:2763), Pontotorula virus (Dalymple et al, 1981, reproduction of New sexual Strand Viruses, Bishop and company (eds.), Elsevier, N.Y., p.167), murine leukemia virus (Steckes et al, 1974, Virol.187), murine virus (Massach virus, S.20), or a hepatitis B virus (hepatitis B virus, or a fragment thereof, for example, british patent publication No. GB 2034323a, published on 04/06/1980; ganem and Varmus,1987, ann.rev.biochem.56: 651693; tiollais et al, 1985, Nature 317: 489495), antigens of equine influenza virus or equine herpes virus (e.g., equine influenza A virus/alaska 91 neuraminidase, equine influenza A virus/miami 63 neuraminidase, equine influenza A virus/kentucky 81 neuraminidase, equine herpes virus glycoprotein B type 1 and equine herpes virus glycoprotein D type I), antigens of bovine respiratory syncytial virus or bovine parainfluenza virus (e.g., bovine respiratory syncytial virus attachment protein (BRSV G), bovine respiratory syncytial virus fusion protein (BRSV F), bovine respiratory syncytial virus nucleocapsid protein (BRSV N), bovine parainfluenza virus fusion protein type 3, bovine parainfluenza virus hemagglutinin neuraminidase type 3), bovine viral diarrhea virus glycoprotein 48 or glycoprotein 53, hepatitis A, influenza, varicella, adenovirus, hepatitis B virus, or herpes virus, Herpes simplex virus type I (HSV I), herpes simplex virus type II (HSV II), rinderpest, rhinovirus, echovirus, rotavirus, respiratory syncytial virus, papilloma virus, pasteurellosis virus, echinoderm virus, arbovirus, hantavirus, coxsackie virus, mumps virus, measles virus, rubella virus, poliovirus, human immunodeficiency virus type II (HIV II), any Picornaviridae, enterovirus, Caliciviridae, any Norwalk virus group, enveloped viruses, such as alphavirus, flavivirus, coronavirus, rabies virus, Marburg virus, Ebola virus, parainfluenza virus, orthomyxovirus, bunyavirus, arenavirus, reovirus, rotavirus, circovirus, human T cell leukemia virus type I, human T cell leukemia virus type II, simian immunodeficiency virus, Lentivirus, polyoma virus, parvovirus, human herpesvirus 6, cervical epithelial herpesvirus I (virus B) and poxvirus.
In certain embodiments, the polypeptide or CAR of the invention binds to HIV, inducing an immune response to the virus in a subject to which the viral vector is administered. Various antigenic domains (e.g., epitopes) of HIV are well known IN the art, and such domains include structural domains, such as Gag, Gag-polymerase, Gag-protease, Reverse Transcriptase (RT), Integrase (IN), and Env. The structural domains of HIV are often further subdivided into polypeptides, e.g., p55, p24, p6 (Gag); p160, p10, p15, p31, p65(pol, prot, RT and IN); gp160, gp120 and gp41(Ems) or Ogp140, as constructed by Chiron Corporation. Molecular variants of such polypeptides may also be targeted for binding by a polypeptide or CAR of the invention, for example, such as those described in PCT/US99/31245, PCT/US99/31273, and PCT/US 99/31272.
Fungi
Examples of fungi (or epitopes thereof) to which a polypeptide or CAR of the invention may bind include, but are not limited to, fungi from the genera Mucor, Candida and Aspergillus, e.g., Mucor racemosus (Mucor leceosus), Candida albicans (Candida albicans) and Aspergillus niger (Aspergillus niger).
Parasite
Examples of parasites (or epitopes thereof) to which a polypeptide or CAR of the invention can bind include, but are not limited to: toxoplasma gondii, Treponema pallidum, Malaria (Malaria), and Cryptosporidium (Cryptosporidium).
Microbial toxin
Examples of microbial toxins (or epitopes thereof) to which a polypeptide or CAR of the invention can bind include, but are not limited to: toxins produced by Bacillus anthracis (Bacillus antrhricus), Bacillus cereus (Bacillus cereus), bordetella pertussis (borfatella pertussis), Clostridium botulinum (Clostridium botulinum), Clostridium perfringens (Clostridium perfringens), Clostridium tetani (Clostridium tetani), Bacillus diphtheriae (corynebacterium dipentiae), Salmonella sp. Such as ricin from canavalia bean and other naturally occurring (e.g., produced by an organism) toxins and artificially produced toxins or portions thereof can also be conjugated to the polypeptides or CARs of the invention.
The pharmaceutical composition may be administered in an appropriate manner to the disease to be treated (or prevented). Although the appropriate dosage can be determined by clinical trials, the amount and frequency of administration will be determined by factors such as the condition of the patient and the type and severity of the patient's disease.
When an "immunologically effective amount", "an effective antitumor amount", "an effective tumor-inhibiting amount", or "therapeutic amount" is indicated, the precise amount of the compound of the present invention to be administered can be determined by a physician, depending on individual differences in age, body weight, tumor size, extent of infection or metastasis, and patient condition (subject). It may be generally noted that 10 may be used4To 109Individual cells/kg body weight (e.g., 10)5To 106Individual cells/kg body weight) of a pharmaceutical composition comprising lymphocytes as described herein, including all integer values within these intervals. The lymphocyte composition may also be administered several times at these doses. It is a general knowledge of immunotherapy that cells can be administered using infusion techniques (see, e.g., Rosenberg et al, New Eng.J.of Med.319:1676,1988). By monitoring the patient for signs of disease and adjusting the treatment accordingly, one skilled in the medical arts can readily determine the optimal dosage and treatment regimen for a particular patient.
Administration of the compositions of the present invention may be carried out in any convenient manner, including infusion or injection (i.e., intravenous, intrathecal, intramuscular, intracavity, intratracheal, intraperitoneal, or subcutaneous), transdermal administration, or other methods known in the art. Administration may be once every two weeks, once a week, or more frequently, but the frequency may be reduced during the disease or condition maintenance period. In some embodiments, the composition is administered by intravenous infusion.
In certain instances, cells activated and expanded using the methods described herein or other methods known in the art, wherein lymphocytes are expanded to therapeutic levels, are administered to a patient (e.g., prior to, concurrently with, or subsequent to) any number of relevant treatment modalities. It is also described herein that lymphocytes can be used in combination with chemotherapy, radiation, immunosuppressive agents, such as cyclosporine, azathioprine, methotrexate, and FK506 antibodies, or other immunoablative agents, such as CAMPATH, anti-cancer antibodies. CD3 or other antibody therapies, cytoxine, fludarabine, cyclosporine, FK506, rapamycin, mycophenolic acid, steroids, FR901228, cytokines and radiation.
The compositions of the invention may also be administered to a patient with (e.g., prior to, concurrently with, or following) bone marrow transplantation, T lymphocyte ablation therapy with chemotherapeutic agents (e.g., fludarabine), radiation external to radiation therapy (XRT), cyclophosphamide, or antibodies (e.g., OKT3 or CAMPATH). It is also described herein that the compositions can be administered after ablative therapy of B lymphocytes (such as agents that react with CD20, e.g., rituximab). For example, a subject may receive standard treatment with high-dose chemotherapy followed by peripheral blood stem cell transplantation. In certain instances, the subject receives an infusion of expanded lymphocytes after transplantation, or the expanded lymphocytes are administered before or after surgery.
In some embodiments, the method may further comprise administering a second therapeutic agent to the subject. The second therapeutic agent is an anti-cancer or anti-neoplastic agent. In some embodiments, the composition is administered to the subject before, after, or simultaneously with the second therapeutic agent (including chemotherapeutic and immunotherapeutic agents).
In some embodiments, the method further comprises administering a therapeutically effective amount of an immune checkpoint modulator. Examples of immune checkpoint modulators may include PD1, PDL1, CTLA4, TIM3, LAG3 and TRAIL. The checkpoint modulator may be administered simultaneously (simultaneousy), separately or concurrently (convurrently) with the composition of the invention.
A "chemotherapeutic agent" is a chemical compound used to treat cancer. Examples of chemotherapeutic agents include alkylating agents such as thiotepa and Cyclophosphamide (CYTOXANTM); alkyl sulfonates such as busulfan, improsulfan and piposulfan; aziridines, such as benzodidopa, carboquinone, methyldopa, and uradopa (uredopa); ethyleneimine and methylmelamine including hexamethylmelamine, triethyleneamineMelamine, triethylenephosphoramide, triethylenethiophosphoramide, and trimethylmelamine; acetogenins (especially bretacine and bretacinone); camptothecin (including the synthetic analog topotecan); bryostatins; statins are cured; CC-1065 (including its synthetic analogs adolesin, kazelesin, bizelesin); nostoc (especially nostoc 1 and nostoc 8); dolastatin; ducamycin (including the synthetic analogs KW-2189 and CBI-TMI); eleutherobin; (ii) coprinus atramentarius alkali; alcohol of coral tree; sponge chalone; nitrogen mustards, such as chlorambucil, chlorphenazine, chlorophosphamide, estramustine, ifosfamide, mechlorethamine, methoxyethylamine hydrochloride, melphalan, neomycin, phenacetin, prednisolone, trofosfamide, uramustine; nitrosoureas, such as carmustine, chlorzotocin, temustine, lomustine, nimustine, ranimustine; antibiotics, such as enediyne antibiotics (e.g., calicheamicin, see, e.g., Agnew chem. Intl. Ed. Engl.33:183-186 (1994); danamicin, including danamicin A; expamicin; and neostatin chromophore and related chromoprotein enediyne antibiotic chromophore), aclacinomycin, actinomycin, amtricin, azaserine, bleomycin, calicheamicin, carubicin, carminomycin, caneomycin, tryptophycetin, dactinomycin, daunorubicin, ditobicin, 6-diazo-5-oxo-L-norleucine, doxorubicin (including morpholino-doxorubicin, cyanomorpholino-doxorubicin, 2-pyrroline-doxorubicin and deoxydoxorubicin), epirubicin, esorubicin, idarubicin, sisomicin, mitomycin, phenolic acid, Streptomycin, oligomycin, pelomomycin, famycin, puromycin, quinomycin, roxobicin, streptomycin, streptozotocin, tubercidin, ubenimex, setastatin, zorubicin; antimetabolites such as methotrexate and 5-fluorouracil (5-FU); analogs such as denopterin, methotrexate, pteropterin, trimetrexate; purine analogs such as fludarabine, 6-mercaptopurine, thioprimine, thioguanine; pyrimidine analogs such as ancitabine, azacitidine, 6-azauridine, carmofur, cytarabine, dideoxyuridine, doxifluridine, enocitabine, fluroureaGlycoside, 5-FU; androgens such as carpoterone, drotaandrosterone propionate, epithioandrostanol, meiandrostane, testolactone; anti-adrenal such as aminoglutethimide, mitotane, trostane; folic acid supplements such as folic acid; acetic acid glucurolactone; an aldehydic phosphoramide glycoside; aminoacetoacetic acid; an acrylic acid ester; the yeast is newer than the yeast; a bisantrene group; edatrexae; 1, ground Budd famine; decarbonylation of colchicine; diazaquinone; eflornithine; ammonium etiolate; an epothilone; etoglut; gallium nitrate; a hydroxyurea; lentinan; lonidamine; maytansinoids, such as maytansine and antomycin; mitoguazone; mitoxantrone; mopidanol; nitraminoacrridine; pentostatin; melphalan; pirarubicin; podophyllinic acid; 2-ethyl hydrazide; procarbazine;lezoxan; lisoxin; west left non-blue; a germanium spiroamine; (ii) zonecanoic acid; a tri-imine quinone; 2, 2' -trichlorotriethylamine; trichodermin (especially T-2 toxin, Veracarin A, rhodamine A and guanidyl pyridine); uratan; vindesine; dacarbazine; mannomustine; dibromomannitol; dibromodulcitol; pipobroman; adding Xituxin; cytarabine ("Ara-C"); cyclophosphamide; thiotepa; taxanes, e.g. paclitaxelAnd docetaxelChlorambucil; gemcitabine; 6-thioguanine; mercaptopurine; methotrexate; platinum analogs, such as cisplatin and carboplatin; vinblastine; platinum; etoposide (VP-16); ifosfamide; (ii) dactinomycin C; mitoxantrone; vincristine; vinorelbine; navelbine, nordstrandon; (ii) teniposide; daunomycin; aminopterin; (ii) Hirodad; ibandronic acid; CPT-11; topoisomerase inhibitor RFS 2000; difluoromethyl ornithine (DMFO); retinoic acid; capecitabine; and pharmaceutically acceptable salts, acids or derivatives of any of the above. Also included in this definition are compounds useful for modulating or inhibiting hormone-mediated processesAnti-hormonal agents of neoplastic action, such as anti-estrogens, including, for example, tamoxifen, raloxifene, aromatase inhibitor 4(5) -imidazole, 4-hydroxytamoxifen, trioxifene, ketoprofen, LY117018, onapristone, and toremifene (faretone); and antiandrogens, such as flutamide, nilutamide, bicalutamide, leuprolide, hiloda, gemcitabine, KRAS mutant covalent inhibitors, and goserelin; and pharmaceutically acceptable salts, acids or derivatives of any of the above. Other examples include irinotecan, oxaliplatin and other standard colon cancer regimens.
An "immunotherapeutic" is a biological agent used to treat cancer. Examples of immunotherapeutics include alemtuzumab, avizumab, bornauzumab, daratumumab, cimizumab, devaluzumab, erlotuzumab, laherparepvec, ipilimumab, nivolumab, atrozumab, ofatumumab, pembrotuzumab, cetuximab, and tammo gene.
In the examples described below, the methods described above were used to enhance the effectiveness of adoptive immunotherapy for cancers expressing the tumor-associated antigen NY-ESO-1 in the context of HLA-A0201 by engineering T cells using TCR-based chimeric antigen receptors that retain high affinity and specificity for NY-ESO-1 in the context of HLA-A0201.
NY-ESO-1 is a protein that is normally expressed only in fetal and testicular tissues, but is aberrantly expressed in some solid malignancies. This adds NY-ESO-1 to a list of molecules that are properly expressed in the germline and are aberrantly expressed by some cancers. These molecules, referred to as "cancer/testis" antigens, can be used as antigen-targeting immunotherapy targets. The cancer-testis antigen NY-ESO-1 is expressed in many solid tumors and is restricted in mature somatic tissues, making it a very attractive target for tumor immunotherapy. Targeting of NY-ESO-1 using engineered T cells has shown clinical efficacy in the treatment of some adult tumors.
The method involves: (1) incorporating a TCR costimulatory signaling element in the CAR design; (2) generating a vector nucleic acid sequence comprising two or more nucleic acid sequences encoding the same polypeptide sequence by silent mutation of one of the exogenous nucleic acid sequences using degenerate codons to reduce homology between the two nucleic acid sequences while maintaining the encoded polypeptide sequence; (3) (ii) integration of a TCR costimulatory signaling element for human CD28, or human 4-1BB, or both; and (4) the same or different transmembrane domains are included in the same TCR-CAR construct. Also disclosed are specific nucleic acid sequences of such CAR genes. This method produces TCR-based CARs with anti-NY-ESO-1 specificity by using the EC domains of the α and β chains of 1G 4195 LY TCR and introducing a combination of TCR signaling element CD3Z or CD3E and co-stimulatory elements CD28, 4-1BB or CD28 and 4-1BB to enhance TCR-CAR expressing T cell activity through TCR-CAR mediated stimulation. This results in an increased antitumor activity in the patient.
Definition of
The T cell receptor or TCR is a complex of proteins present on the surface of T cells or T lymphocytes, which is responsible for recognizing antigen fragments as peptides bound to MHC molecules. The binding between the TCR and the antigenic peptide is of relatively low affinity and is degenerate; that is, many TCRs recognize the same antigenic peptide and many antigenic peptides are recognized by the same TCR. Most T cells have TCRs in the form of multiple protein complexes. The TCR consists of two distinct protein chains. In humans, the TCR consists of α (α) and β (β) chains (encoded by TRA and TRB, respectively) in 95% of T cells, whereas the TCR consists of γ and δ (γ/δ) chains (encoded by TRG and TRD, respectively) in 5% of T cells. This ratio varies during development and under diseased conditions (e.g., leukemia). Also vary between species. Each chain of the TCR consists of two extracellular domains: a variable (V) region and a constant (C) region. The constant region is near the cell membrane, followed by a transmembrane region and a short cytoplasmic tail, while the variable region binds to the peptide/MHC complex. For the purposes of the present invention, the term "constant region of a T cell receptor chain or a portion thereof" also includes embodiments in which the constant region of a T cell receptor chain (from N-terminus to C-terminus) is followed by a transmembrane region and a cytoplasmic tail, such that the transmembrane region and cytoplasmic tail are naturally linked to the constant region of the T cell receptor chain.
As used herein, the term "antigen receptor" or "antigen recognition receptor" refers to a receptor that is capable of activating an immune cell (e.g., a T cell) in response to antigen binding. In particular, the term "antigen receptor" includes engineered receptors that confer any specificity to immune effector cells (e.g., T cells). The antigen receptor according to the invention may be present on a T cell, e.g. instead of or in addition to the T cell's own T cell receptor. Such T cells do not necessarily need to process and present antigens in order to recognize target cells, but may preferably specifically recognize any antigen present on target cells. Preferably, the antigen receptor is expressed on the surface of a cell. In particular, the term includes artificial or recombinant receptors comprising a single molecule or complex of molecules that recognizes (i.e., binds to) a target structure (e.g., an antigen) on a target cell (e.g., by binding an antigen binding site or antigen binding domain to an antigen expressed on the surface of the target cell) and can confer specificity for an immune effector cell (e.g., a T cell) that expresses the antigen receptor on the surface of the cell. Preferably, recognition of the target structure by the antigen receptor results in activation of an immune effector cell expressing the antigen receptor. The antigen receptor may comprise one or more protein units comprising one or more domains as described herein. The term "antigen receptor" preferably does not include naturally occurring T cell receptors. According to the present invention, the term "antigen receptor" is preferably synonymous with the terms "chimeric antigen receptor", "chimeric T cell receptor" and "artificial T cell receptor".
Exemplary antigen recognizing receptors can be native or genetically engineered TCRs, or genetically engineered TCR-like mabs (Hoydahl et al, Antibodies 20198: 32) or CARs, in which a tumor antigen binding domain is fused to an intracellular signaling domain capable of activating an immune cell (e.g., a T cell). T cell clones expressing native TCRs against specific Cancer Antigens have been previously disclosed (Traversari et al, J Exp Med, 1992176: 1453-7; Ottavalini et al, Cancer Immunol Immunother, 200554: 1214-20; Chaux et al, J Immunol, 1999163: 2928-36; Luiten and van der Bruggen, Tissue antibodies, 200055: 149-52; van der Bruggen et al, Eur J Immunol, 567: 3038-43; Huang et al, J Immunol, 1999162: 6849-54; Ma et al, Int J Cancer, 2004109: 698-, Ebert et al, Cancer Res, 200969: 1046-54; Ayub et al, J17128: 1717-22; Chamunko et al, Conn Immunol et al, Watson et al, J Immunol, 200 201060: 200756; Journal of beer et al, Cancer J Bruggen, J Bruggen et al, J Bruggen, Gn J Bruggen, J Bruggen et al, J Bruggen, J Brugen et al, J Brugen, 2004109: 698-702, J Brugen, PNAS, 2003100: 8862-67; chen et al, PNAS, 2004).
The term "chimeric antigen receptor" or "CAR" refers to a group of polypeptides, usually two in the simplest embodiment, which, when in an immune effector cell, provide a cell that is specific for the target cell and produces an intracellular signal. In some embodiments, the CAR comprises at least an extracellular antigen-binding domain, a transmembrane domain, and a cytoplasmic signaling domain (also referred to herein as an "intracellular signaling domain") that comprises a functional signaling domain derived from a stimulatory molecule and/or a co-stimulatory molecule as defined below. In some embodiments, the set of polypeptides are in the same polypeptide chain, e.g., comprise a chimeric fusion protein. In some embodiments, the set of peptides are discontinuous with respect to each other, e.g., in different polypeptide chains. In some embodiments, the set of polypeptides comprises a dimerization switch that is capable of coupling the polypeptides to each other, e.g., capable of coupling an antigen binding domain to an intracellular signaling domain, when a dimerization molecule is present. In one aspect, the co-stimulatory molecule of the CAR is a zeta chain that binds to the T cell receptor complex (e.g., CD3 zeta). In one aspect, the cytoplasmic signaling domain comprises a primary signaling domain (e.g., the primary signaling domain of CD 3-zeta). In one aspect, the cytoplasmic signaling domain further comprises one or more functional domains derived from at least one co-stimulatory molecule as defined below. In one aspect, the co-stimulatory molecule is selected from the co-stimulatory molecules described herein, e.g., 4-1BB (i.e., CD137), CD27, and/or CD 28.
The term "TCR-based CAR" or "TCR-CAR" refers to a CAR that comprises an antigen binding domain, or antigen binding portion thereof, formed by TCR alpha, beta, gamma, or delta chains.
The term "stimulatory molecule" refers to a molecule expressed by an immune cell (e.g., a T cell, NK cell, or B cell) that provides one or more cytoplasmic signaling sequences that modulate immune cell activation in a stimulatory manner with respect to at least some aspects of the immune cell signaling pathway. In one aspect, the signal is the primary signal, e.g., initiated by binding of the TCR/CD3 complex to peptide-loaded MHC molecules, and results in the mediation of T cell responses, including, but not limited to, proliferation, activation, differentiation, and the like. The major cytoplasmic signaling sequence (also referred to as the "major signaling domain") that functions in a stimulatory manner may contain signaling motifs referred to as immunoreceptor tyrosine-based activation motifs or ITAMs. Examples of ITAM-containing cytoplasmic signaling sequences particularly useful in the present invention include, but are not limited to, those derived from CD3 ζ, common FcR γ (FCER1G), fcyriia, FcR β (fcepsilonr 1b), CD3 γ, CD3 Δ, CD3 ∈, CD79a, CD79b, DAP10, and DAP 12. In a specific CAR of the invention, the intracellular signaling domain in any one or more CARs of the invention comprises an intracellular signaling sequence, e.g., the major signaling sequence of CD 3-zeta. In specific CARs of the invention, the primary signaling sequence for CD 3-zeta is the sequence provided herein, or equivalent residues from non-human species, e.g., mouse, rodent, monkey, ape, etc. In specific CARs of the invention, the primary signaling sequence for CD 3-zeta is a sequence as provided herein, or equivalent residues from a non-human species, e.g., mouse, rodent, monkey, ape, etc.
The term "co-stimulatory molecule" refers to a cognate binding partner that specifically binds to a co-stimulatory ligand on a T cell, thereby mediating a co-stimulatory response of the T cell, such as, but not limited to, proliferation. Costimulatory molecules are cell surface molecules other than antigen receptors or their ligands, which contribute to an effective immune response. Costimulatory molecules include, but are not limited to, MHC class I molecules, TNF receptor proteins, immunoglobulin-like proteins, cytokine receptors, integrins, signaling lymphocyte activating molecules (SLAM proteins), activated NK cell receptors, BTLA, Toll ligand receptors, OX40, CD2, CD7, CD27, CD28, CD30, CD40, CDS, ICAM-1, LFA-1(CD11a/CD18), 4-1BB/CD137, B7-H7, CDS, ICAM-1, ICOS (CD278), GITR, BAFFR, LIGHT, HVEM (LIGHT TR), KIRDS 7, SLAMF7, NKp 7 (KLRF 7), NKp 7, CD7 β, IL2 γ, VLITGA 7, GAITGB 7, GAITGA 7, GAITGB 11, GAITGA 7, GAITGB 7, GAIT11-7, GAITGB 7, GAIT11, GAITGA 7, GAITGB 7, CD7, GAIT11, CD7, GAITGB 7, GAIT11-7, GAITGB 7, GAIT11, TRANCE/RANKL, DNAM1(CD226), SLAMF4(CD244, 2B4), CD84, CD96 (tactle), CEACAM1, CRTAM, Ly9(CD229), CD160(BY55), PSGL1, CD100(SEMA4D), CD69, SLAMF6(NTB-A, Ly108), SLAM (SLAMF1, CD150, IPO-3), BLAME (SLAMF8), SELPLG (CD162), LTBR, LAT, GADS, SLP-76, PAG/Cbp, CD19a, and a ligand that specifically binds to CD 83.
The costimulatory intracellular signaling domain refers to the intracellular portion of the costimulatory molecule. The intracellular signaling domain may comprise the entire intracellular portion of the molecule from which it is derived or the entire native intracellular signaling domain, or a functional fragment or derivative thereof.
As used herein, two polypeptide (or nucleic acid) sequences are "substantially different" meaning that the two sequences are less than 95% (e.g., 90%, 85%, 80%, 75%, 70%, 65%, 60%, 55%, or 50%) identical to each other.
As used herein, the term "functional variant" refers to a modified polypeptide or protein or transgene having substantial or significant sequence identity or similarity to the wild-type, such functional variant retaining the biological activity of the wild-type polypeptide or protein or transgene for which it is a variant. In some embodiments, a functional variant of a therapeutic polypeptide or protein or transgene is used.
Conservative modifications or functional equivalents of the peptides, polypeptides or proteins disclosed herein refer to polypeptide derivatives of the peptides, polypeptides or proteins, e.g., proteins having one or more point mutations, insertions, deletions, truncations, fusion proteins, or combinations thereof. Which substantially retains the activity of the parent peptide, polypeptide or protein (such as those disclosed in the present invention). In general, a conservative modification or functional equivalent is at least 60% (e.g., any number between 60% and 100%, including, e.g., 60%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, and 99%) identical to the parent (e.g., one of those disclosed herein). Thus, within the scope of the present invention, the hinge region has one or more point mutations, insertions, deletions, truncations, fusion proteins, or combinations thereof.
In some cases, the structure of the peptide backbone in the region of the substitution (1) is maintained, (b) the charge or hydrophobicity of the molecule at the target site; or (c) substitution in which the effect is not significantly different in side chain volume. For example, naturally occurring residues can be classified into several classes based on the nature of the side chains: (1) hydrophobic amino acids (norleucine, methionine, alanine, valine, and isoleucine); (2) neutral hydrophilic amino acids (cysteine, serine, threonine, asparagine, and glutamine); (3) acidic amino acids (aspartic acid and glutamic acid); (4) basic amino acids (histidine, lysine and arginine); (5) amino acids that affect chain orientation (glycine and proline); and (6) aromatic amino acids (tryptophan, tyrosine, and phenylalanine). Substitutions made in these categories may be considered conservative substitutions. Examples of substitutions include, but are not limited to, valine for alanine, lysine for arginine, glutamine for asparagine, glutamic acid for aspartic acid, serine for cysteine, asparagine for glutamine, aspartic acid for glutamic acid, proline for glycine, arginine for histidine, leucine for isoleucine, isoleucine for leucine, arginine for lysine, leucine for methionine, leucine for phenylalanine, glycine for proline, threonine for serine, serine for threonine, tyrosine for tryptophan, phenylalanine for tyrosine, and/or leucine for valine. Exemplary substitutions are shown in the table below. Amino acid substitutions can be introduced into the parent protein and the product screened for retention of the biological activity of the parent protein.
Original residues
Exemplary substitutions
Ala(A)
Val;Leu;Ile
Arg(R)
Lys;Gln;Asn
Asn(N)
Gln;His;Asp,Lys;Arg
Asp(D)
Glu;Asn
Cys(C)
Ser;Ala
Gln(Q)
Asn;Glu
Glu(E)
Asp;Gln
Gly(G)
Ala
His(H)
Asn;Gln;Lys;Arg
Ile(I)
Leu; val; met; ala; phe; norleucine
Leu(L)
Norleucine; ile; val; met; ala; phe (Phe)
Lys(K)
Arg;Gln;Asn
Met(M)
Leu;Phe;Ile
Phe(F)
Trp;Leu;Val;Ile;Ala;Tyr
Pro(P)
Ala
Ser(S)
Thr
Thr(T)
Val;Ser
Trp(W)
Tyr;Phe
Tyr(Y)
Trp;Phe;Thr;Ser
Val(V)
Ile; leu; met; phe; ala; norleucine
As used herein, the percent homology between two amino acid sequences is equal to the percent identity between the two sequences. The percent identity between two sequences is a function of the number of identical positions shared by the sequences (i.e.,% homology-number of identical positions/total number of positions x 100), where the number of gaps, and the length of each gap, that need to be introduced to achieve optimal alignment of the two sequences, are taken into account. As described in the following non-limiting examples, comparison of sequences and determination of percent identity between two sequences can be accomplished using a mathematical algorithm.
The percent identity between two amino acid sequences can be determined using the e.meyers and w.miller algorithms (comput.appl.biosci.,4:11-17(1988)), which have been integrated into the ALIGN program (version 2.0) using a PAM120 residue weight table, a gap length penalty of 12 and a gap penalty of 4. Furthermore, the percent identity between two amino acid sequences can be determined using the Needleman and Wunsch (J.mol.biol.48:444-453(1970)) algorithm, which has been integrated into the GAP program in the GCG software package (available from www.gcg.com), using either the Blossum 62 matrix or the PAM250 matrix, the GAP weights 16, 14, 12, 10, 8,6, or 4 and the length weights 1, 2, 3, 4,5, or 6.
As used herein, the term "antibody" refers not only to an intact antibody molecule, but also to fragments of an antibody molecule that retain the ability to bind antigen. Such fragments are also well known in the art and are used regularly, both in vitro and in vivo. Thus, as used herein, the term "antibody" refers not only to intact immunoglobulin molecules, but also to the well-known active fragments f (ab') 2 and fab. F (ab') 2 and fab fragments lacking the Fe fragment of the intact antibody are cleared more rapidly from the circulation and are less likely to bind to non-specific tissues of the intact antibody (Wahl et al, J.Nucl. Med.24:316-325 (1983)). Antibodies of the invention include intact natural antibodies, bispecific antibodies; a chimeric antibody; fab, fab', single chain v region fragments, fusion polypeptides, and non-canonical antibodies.
As used herein, the term "single-chain variable fragment" or "scFv" is a fusion protein of the variable regions of the heavy (VH) and light (VL) chains of an immunoglobulin covalently linked to form a VH:: VL heterodimer. The heavy chain (VH) and light chain (VL) are linked directly or through a peptide-encoding linker (e.g., 10, 15, 20, 25 amino acids) that links the N-terminus of the VH to the C-terminus of the VL, or the C-terminus of the VH to the N-terminus of the VL. The linker is typically rich in glycine to improve flexibility and serine or threonine to improve solubility. Despite the removal of the constant region and the introduction of the linker, the scFv protein retains the specificity of the original immunoglobulin. Single chain Fv polypeptide antibodies can be expressed from nucleic acids comprising VH and VL coding sequences as described by Huston et al (Proc. Nat. Acad. Sci.,85:5879-5883, 1988). See also, U.S. Pat. nos. 5,091,513, 5,132,405, and 4,956,778; and U.S. patent publication nos. 20050196754 and 20050196754. Antagonistic scFvs with inhibitory activity have been described (see, e.g., Zhao et al, hybridoma (Larchmont) 200827 (6): 455-51; Peter et al, Jcachexia saropenia muscle 2012 Aug.12; Shieh et al, J Immunol 2009183(4): 2277-85; Giomarelli et al, Thromb Haemost 200797 (6): 955-63; Fife et al, J Clin Invst 2006116 (8): 2252-61; Brocks et al, Immunol technology 19973(3): 173-84; Moosmayer et al, Immunol 19952 (10: 31-40)). Agonistic scFv with stimulatory activity have been described (see, e.g., Peter et al, J Bio Chern 200325278(38): 36740-7; Xie et al, Nat Biotech 199715 (8): 768-71; Ledbetter et al, Crit Rev Immunol 199717 (5-6): 427-55; Ho et al, Biochim Biophys Acta 20031638(3): 257-66).
As used herein, "treating" or "treatment" refers to administering a compound or agent to a subject having a disorder for the purpose of curing, alleviating, remedying, delaying the onset of, preventing, or ameliorating the disorder, a symptom of the disorder, a disease state secondary to the disorder, or a susceptibility to the disorder.
An "effective amount" or "therapeutically effective amount" refers to the amount of a compound or agent that is capable of producing a medically desirable result in a treated subject. The treatment may be performed in vivo or ex vivo, alone or in combination with other drugs or therapies. A therapeutically effective amount may be administered in one or more administrations, applications or dosages and is not intended to be limited to a particular formulation or route of administration.
The terms "effective amount", "effective dose" or "effective dose" are defined as an amount sufficient to achieve, or at least partially achieve, the desired effect. By "therapeutically effective amount" or "therapeutically effective dose" of a drug or therapeutic agent is meant any amount of drug that, when used alone or in combination with another therapeutic agent, promotes disease regression as manifested by decreased severity of disease symptoms, increased frequency and duration of disease symptom-free periods, or prevention of dysfunction or disability due to disease affliction. A "prophylactically effective amount" or "prophylactically effective dose" of a drug is an amount of the drug that, when administered alone or in combination with another therapeutic agent, is administered to a subject at risk of developing a disease or experiencing a relapse of the disease, to inhibit the development or relapse of the disease. The ability of a therapeutic or prophylactic agent to promote regression of a disease or inhibit the progression or recurrence of said disease can be assessed using a variety of methods well known to the skilled artisan, such as in human subjects during clinical trials, in animal model systems predicting efficacy in humans, or by assaying the activity of the agent in an in vitro assay.
As used herein, the term "pharmaceutically acceptable" refers to a substance (e.g., carrier or diluent) that does not eliminate the biological activity or properties of the composition and is relatively non-toxic, i.e., the substance can be administered to an individual without causing deleterious biological effects or interacting in a deleterious manner with any of the components of the composition contained therein.
The term "pharmaceutically acceptable carrier" includes pharmaceutically acceptable salts, pharmaceutically acceptable substances, compositions or vehicles, such as liquid or solid fillers, diluents, excipients, solvents or encapsulating substances, which are involved in carrying or transporting one or more compounds of the invention within or to a subject such that they may perform their intended function. Typically, such compounds are carried or transported from one organ or part of the body to another organ or part of the body. Each salt or carrier must be "acceptable" in the sense of being compatible with the formulation and other ingredients, and not deleterious to the subject. Some examples of substances that may serve as pharmaceutically acceptable carriers include: sugars such as lactose, glucose and sucrose; starches, such as corn starch and potato starch; cellulose and its derivatives, such as sodium carboxymethyl cellulose, ethyl cellulose and cellulose acetate; powdered tragacanth; malt; gelatin; talc powder; excipients, such as cocoa butter and suppository waxes; oils such as peanut oil, cottonseed oil, safflower oil, sesame oil, olive oil, corn oil and soybean oil; glycols, such as propylene glycol; polyols such as glycerol, sorbitol, mannitol and polyethylene glycol; esters, such as ethyl oleate and ethyl laurate; agar; buffering agents such as magnesium hydroxide and aluminum hydroxide; alginic acid; pyrogen-free water; isotonic saline; ringer's solution; ethanol; phosphate buffered saline; a diluent; granulating; a lubricant; a binder; a disintegrant; a wetting agent; an emulsifier; a colorant; a release agent; a coating agent; a sweetener; a flavoring agent; a flavoring agent; a preservative; an antioxidant; a plasticizer; a thickener; a thickener; a hardening agent; a curing agent; a suspending agent; a surfactant; a humectant; a carrier; a stabilizer; and other non-toxic compatible materials used in pharmaceutical formulations, or any combination thereof. As used herein, "pharmaceutically acceptable carrier" also includes any and all coatings, antibacterial and antifungal agents, and absorption delaying agents, and the like, that are compatible with the activity of the compound and are physiologically acceptable to a subject. Supplementary active compounds may also be added to the composition.
As used herein, the term "about" or "approximately" when applied to one or more values refers to a value similar to the stated reference value. In some embodiments, the term "about" or "approximately" refers to a range of values that fall within 25%, 20%, 19%, 18%, 17%, 16%, 15%, 14%, 13%, 12%, 11%, 10%, 9%, 8%, 7%, 6%, 5%, 4%, 3%, 2%, 1%, or less, in either direction (greater or less) of a stated reference value, unless otherwise stated or apparent from the context (unless the number therein would exceed 100% of the possible values). Unless otherwise indicated herein, the term "about" is intended to include values (e.g., weight percentages) near the stated range that are functionally equivalent in terms of individual ingredients, compositions, or embodiments. It is to be understood that wherever values and ranges are provided herein, all values and ranges subsumed by such data and ranges are intended to be encompassed within the scope of the present invention. Further, the present application also includes all values falling within these ranges and the upper or lower limits of the ranges of values.
Examples
Example 1: construction of MGF retroviral vector based on NY-ESO-1TCR-, NY-ESO-1TCR-CAR-, mesothelin-and GFP-resistant
Construction of retroviral constructs
In order to constitutively express a target gene in T cells, recombinant retroviral vectors encoding the target gene, such as TCR (NT1, NT1B) or TCR-CAR (NT2, NT3, NT4, NT5, NT6, NT21, NT22, NT23, NT24, NT25, NT26, NT27 and T28) specifically directed against the native form of the human NY-ESO-1 derived peptide/a 2 complex were originally constructed using mfg. MFG retroviral backbone and SFG retroviral backbone (us patent 6,140,111) developed by Mulligan doctor (fig. 1C) (fig. 1A and fig. 1B and table 1).
Table 1: list of genes encoding anti-NY-ESO-1 TCR, anti-NY-ESO-1 TCR-CAR, anti-mesothelin CAR and GFP
GFP was used as a marker to track T cells transduced by the GFP-based vector by flow cytometry (FACS) and/or by fluorescence microscopy. Anti-human mesothelin sFv (GenBank ID: AF035617.1) (Chowdhury PS et al, Proc Natl Acad Sci U S A.1998; 95:669-74) with high affinity for human mesothelin was selected for construction of anti-mesothelin based CARs. This use of anti-mesothelin sFv has been described previously in the successful establishment of second and third generation functional anti-mesothelin CARs (Carpentio C. et al, Proc Natl Acad Sci U S A.2009; 106: 3360-5).
Recombination between nucleic acids is a well-recognized phenomenon in molecular biology. Genetic recombination, which requires strong sequence homology between the participating nucleic acid sequences, is generally referred to as homologous recombination. Although most gene knockout strategies employ homologous recombination to achieve targeted knockouts, in some systems, the occurrence of gene recombination can adversely affect gene manipulation. In particular, homologous recombination events can adversely affect the construction and production of vectors, particularly viral vectors (e.g., adenoviruses, retroviruses, adeno-associated viruses, herpes viruses, and the like), where it is often desirable to maintain highly homologous sequences (e.g., identical polypeptide sequences) within a single, stable viral vector without homologous recombination during, for example, passage and/or propagation of the viral vector through one or more host cells and/or organisms.
The methods described previously (U.S. patent No. 9,206,440) were used in vector design to overcome such problems associated with potential homologous recombination between nucleic acids to enable delivery of two or more nucleic acid sequences encoding highly homologous (e.g., identical) polypeptide fragments in a single protein molecule on a single viral vector. In this method, silent mutations at the nucleotide sequence level are employed to generate viral vector sequences comprising nucleic acid sequences encoding two or more highly homologous (e.g., identical) polypeptides or polypeptide domains thereof, but with a reduced risk of homologous recombination between such nucleic acid sequences, even in, for example, prolonged passages and/or multiple infections, chromosomal integration and/or excision events in a host cell.
Mutant TCRs of peptide SLLMWITQC (SEQ ID NO:25) corresponding to residues 157 to 165 of NY-ESO-1 (NY-ESO-1:157-165) have been previously described (Robbins PF et al, J Clin Oncol.2011; 29: 917-24; and Robbins PF et al, J Immunol.2008; 180: 6116-6131) recognizing with high affinity in the context of HLA-A0201 class I restriction elements. This TCR, designated 1G4- α 95LY, comprises two amino acid substitutions in the third complementarity determining region of the native 1G4TCR α chain, which confers enhanced ability of CD8+ and CD4+ T cells to recognize HLA-a 0201-positive target cells expressing the NY-ESO-1 antigen. The entire alpha and beta chain of TCR IG4-a95LY (SEQ ID NO:1 and SEQ ID NO:2) or an extracellular domain thereof (SEQ ID NO:4 and SEQ ID NO:5) two anti-NY-ESO-1 TCRs (NT1, NT1b) used to generate native form IG4-a95LY TCR (SEQ ID NO:9,64) or two TCR-CARs (IG4-a95LY TCR-CARs) (NT 2; IG4-a95LY TCR-based CAR-CD28Z (aNY-b/a28tm28Z), (SEQ ID NO:10), and NT3, IG4-a95LY TCR-based CAR-CD28E (aNY-b/a28tm28E) (SEQ ID NO:11) (FIGS. 1 and 2) of the drawings, which recognizes with high affinity the peptide SLLMWITQC (SEQ ID NO: 25).
The MGF retroviral backbone mentioned above was used to construct recombinant viral vectors encoding one of the two anti-NY-ESO-1 TCRs in native form (vector 1 and vector 2; FIGS. 1A, 1B), one of the twelve anti-NY-ESO-1 TCR based CARs (vectors 113-15; FIGS. 1A, 1B) or anti-human mesothelin-CAR or GFP.
(1) Native form of 1G4-a95LY TCR-vector against NY-ESO-1/A2 (FIG. 1A, FIG. 1B, FIG. 2A, FIG. 2B):
carrier 1(NT1, NT1a, aNY-TCRa/b) (SEQ ID NOS: 9 and 12)
MFG-based retroviral vectors encoding the TCR recognizing the peptide SLLMWITQC (SEQ ID NO:25) in the context of HLA-A0201 class I restriction elements were generated in the retroviral vector backbone as described previously (Robbins PF et al, J Clin Oncol.2011; 29:917-24 and Robbins et al, J Immunol.2008; 180: 6116-6131). This TCR, designated 1G4- α 95: LY, contains two amino acid substitutions in the third complementarity determining region of the native 1G4TCR α chain, which confers enhanced ability of CD8+ and CD4+ T cells to recognize HLA-A0201-positive target cells expressing the NY-ESO-1 antigen (Robbins PF et al, J Immunol.2008; 180: 6116-.
The 1G4 alpha-and beta-strands expressed in the retroviral construct contained a "self-cleaving" P2A sequence between the two gene products (Szymczak et al, nat. Biotechnol. 2004; 22: 589. sup. 594). The peptide sequence (aNY-TCRa/b) (SEQ ID NO:9) (FIG. 3) of NT1 was designed to be encoded by the nucleotide sequence (SEQ ID NO: 12). To facilitate molecular subcloning of the target gene in the cloning site of the MFG-based vector, an XhoI site (CTCGAG, SEQ ID NO:71) and a short sequence (CAGCCAGCGGCCGC, SEQ ID NO:72) comprising a NotI site (GCGGCCGC, SEQ ID NO:73) were inserted immediately upstream of the start codon of "ATG" and downstream of the stop codon "TAA" (SEQ ID NO:12), respectively, in the coding region.
Carrier 2(NT1 b; aNY-TCRb/a)
The vector (NT 2; aNY-TCRb/a) (SEQ ID NOS: 64 and 65) was designed identically to vector 1(NT 1; aNY-TCRa/b), except that the DNA fragment encoding the native form of the TCR α chain was located immediately downstream of the 3' end of the P2A coding sequence, while the β chain was located upstream of the P2A coding sequence (FIGS. 1, 2 and 3).
(2) 1G4-a95LY TCR-CAR-vectors (NT2, NT3) integrated with CD28 and CD3Z signaling (FIGS. 1C and 1D)
Carrier 3(NT 2; aNY-b/a28tm28Z) (second generation)
Recombinant retroviruses encoding two or more molecules that are identical or highly homologous in nucleic acid sequence may result in homologous recombination and, as a result, genomic rearrangements, such as deletions and duplications of the homologous genes, may occur. For example, during plasmid DNA and retroviral vector production, nucleic acid sequences encoding the Tm and Cyt domains of TCR-based CARs (e.g., CD28TmCyt, CD3ZCyt, and CD3ECyt segments) may result in undesirable mutations and/or deletions of the gene encoding TCR-CAR. To address this problem, mutation programs can be used to suppress homology-driven recombination between repeated segments in the same vector.
The amino acid GSPK (SEQ ID NO:69) preceding the 28Z or 28E peptide sequence was used as a linker (FIGS. 4 and 5) and linked to TCRa (at the TCR-Ca amino acid end of SPESS) and TCRb (at the TCR-Cb amino acid end of WGRAD) as described previously (Govers et al, Journal of Immunology,2014,193: 5315-5326).
NT2 (vector 3; aNY-b/a28tm28Z) (1G 4a 95LY TCR based CAR) was designed to contain 28Z signaling (FIGS. 1C and 2C) (SEQ ID NO: 10).
To construct NT2 (FIG. 2C; SEQ ID NO:10, 13), the extracellular domain of the beta chain (SEQ ID NO:5) and the extracellular domain of the alpha chain (SEQ ID NO:4) were linked via linkers (amino acid GSPK (SEQ ID NO:69)) to Tm and Cyt (28TmCyt) (SEQ ID NO:6) of human CD28 and Cyt (ZCyt) (SEQ ID NO:7) molecules of human CD3Z, wherein CD28Z (SEQ ID NO:15) directly linked to the extracellular domain of the 1G 4a 95LY TCR alpha chain was silent mutated without changing its original amino acid sequence (mu28muZ) (FIGS. 6-8) (SEQ ID NO:21) to reduce homology at the level of nucleotide sequences encoding 28Z between those sequences directly linked to the C-termini of the beta and alpha chains Ec. Similar to the native form of 1G 4a 95LY TCR (NT1 and NT1b), a "self-cleaving" P2A sequence (SEQ ID NO:3) was also inserted between β CD28Z and α CD28muZ (FIG. 4).
Carrier 13(NT 26; aNY-b/a8h28pectm28Z) (second generation)
Vector 13(NT26, SEQ ID NOS: 40 and 62) was designed and prepared in the same manner as vector 3(NT2) except that two DNA sequences encoding the same amino acid sequences of the human CD8a hinge (CD8h) (SEQ ID NO:30) and a part of the human CD28 extracellular domain (CD28pec) (SEQ ID NO:26) were inserted between amino acids GSPK (SEQ ID NO:69) linked to the C-terminus of Cb and the N-terminus (SEQ ID NO:42,50) of CD28Tm and between amino acids GSPK (SEQ ID NO:69) linked to the C-terminus of Ca and the N-terminus (SEQ ID NO:43,51) of CD28 Tm.
(3) 1G4-a95LY TCR-CAR-vector integrated with CD28 and CD3E signaling (NT 3; aNY-b/a28tm28E) (FIGS. 1 and 2)
Carrier 4(NT 3; aNY-b/a28tm28E) (second generation)
The 28E expression cassette was designed at the amino acid sequence level as described previously, covering the Tm and Cyt (IC) domains of human CD28 (GI: 338444, aa 153-. The 28 ε expression cassette is preceded by the amino acid GSPK (SEQ ID NO:69) and is linked downstream of the Ec (TCR-Ca amino acids ending in SPESS) and TCRb (TCR-Cb amino acids ending in WGRAD) of the C α region. A nucleotide sequence encoding NT3(SEQ ID NO:14) was designed which contained the nucleotide sequence of Xho I immediately upstream of the coding region at the aNY-TCR28E site, as well as a short fragment flanking the 3 major terminus and a Not I site (FIGS. 2D, 5 and 11) (SEQ ID NO:14) (SEQ ID NO:11) and was synthesized commercially (BIO BASIC CANADA INC, Canada). G4 a95LY TCR based CAR comprises 28E signaling (NT 3; 1G 4a 95LY TCR-28E) (aNY-b/a28tm28E) (FIGS. 1D and 2D).
Similar to aNY-TCR28Z (NT 2; vector 3), to construct an IG 4a 95LY TCR-based CAR comprising 28E signaling (1G 4a 95LY TCR-28E; aNY-TCR 28E; vector 3), the Ec domains of the β and α chains were linked via a linker (amino acid GSPK (SEQ ID NO:69)) to the Tm and Cyt domains of human CD28 and the Cyt molecule of human CD3E, with CD28E linked directly to the extracellular domain of the 1G 4a 95LY TCR α chain being silently mutated without altering its original amino acid sequence (mu28muE) (fig. 6) to reduce homology at the level of nucleotide sequences encoding 28E between those coding for direct linkage to the Ec termini of the β and α chains. Similar to 1G 4a 95LY TCR, a "self-cleaving" P2A sequence (SEQ ID NO:3) was also inserted between β CD28E and α muCD28muE (FIG. 5).
(4) 1G4-a95LY TCR-CAR-vector (vector 5(NT 4; aNY-b/a8tmBBZ) and vector 6(NT 5; aNY-b/a28tmBBZ)) integrated with 4-1BB and CD3Z signaling
Carrier 5(NT 4; aNY-b/a8tmBBZ) (second generation)
Vector 5(SEQ ID NOS: 32 and 54) was designed and prepared in the same manner as vector 3(NT2) except that the DNA sequences encoding the Tm and Cyt domains of CD28 were replaced with DNA sequences encoding the Tm domain of human CD 8(SEQ ID NOS: 31) (SEQ ID NOS: 52 and 53) and the Cyt domain of human 4-1BB (SEQ ID NOS: 29) (SEQ ID NOS: 48, 49).
Carrier 6(NT 5; aNY-b/a28tmBBZ) (second generation)
Vector 6(NT 5; aNY-b/a28tmBBZ) (SEQ ID NOS: 33,55) was designed and prepared in the same manner as vector 5(NT4) except that the DNA sequence encoding the Tm domain of CD8 was replaced with the DNA sequence encoding the Tm domain of human CD28 (SEQ ID NOS: 29) (SEQ ID NOS: 48 and 49).
(5) 1G4-a95LY TCR-CAR-vector integrated with a combination of CD28 and 4-1BB and CD3Z signaling (vector 7(NT6), vector 8(NT21), vector 9(NT22), vector 10(NT23), vector 11(NT24), vector 12(NT25), vector 14(NT27))
Carrier 7(NT 6; aNY-b/a28tm28BBZ) (third generation)
Vector 7(SEQ ID NO:34,56) was designed and prepared in the same manner as vector 3(NT2) except that the DNA sequences encoding the Cyt domain of human 4-1BB (SEQ ID NO:29) (SEQ ID NOS: 48 and 49) were inserted between the C-terminal of CD28 and the N-terminal Cyt domain of CD3Z, respectively.
Carrier 11(NT 24; aNY-b8tm28Z/a8tmBBZ) (third generation)
Vector 11(SEQ ID NO:38,60) was designed and prepared in the same manner as vector 5(NT 4; aNY-b/a8tmBBZ), except that the DNA sequence encoding the Cyt domain of 4-1BB, located between the C-terminus of CD8Tm and the N-terminus of CD3Zcyt in the first polypeptide comprising the Ec domain of the TCR β chain (i.e., upstream of the P2A peptide), was replaced with the DNA sequence encoding the Cyt domain of human CD 28.
Carrier 12(NT 25; aNY-b8tm2828Z/a8 tmBBBBBBZ) (third generation)
Vector 12(SEQ ID NO:39 and 61) was designed and prepared in the same manner as vector 11(NT24) except that an additional DNA sequence encoding the Cyt domain of CD28 (unit 2 of CD28 Cyt) was inserted between the C-terminus of CD8Tm and the N-terminus of CD29Cyt in the first polypeptide chain containing Ec against the NY-ESO-1TCR β chain, and an additional DNA sequence encoding the Cyt domain of 4-1BB (unit 2 of 4-1 BBcyt) was inserted between the C-terminus of CD8Tm and the N-terminus of 4-1BB in the second polypeptide chain containing Ec against the NY-ESO-1TCR α chain.
Accordingly, molecular design and methods similar to those described above were used to design and prepare other vectors, including vector 8(SEQ ID NO:35, 57; NT 21; aNY-b28tm28Z/a8tmBBZ), vector 9(SEQ ID NO:36, 58; NT 22; aNY-b28tm28Z/a8tmBB), vector 10(SEQ ID NO:37, 59; NT 23; aNY-b28tm28Z/a28tmBBZ), vector 14(SEQ ID NO:41, 63; NT 24; aNY-b8h28pectm28Z/a8tmBB) (Table 1).
DNA fragments encoding 14 anti-NY-ESO-1 TCR or TCR-CAR (vectors 1-14; Table 1) were synthesized commercially (BIO BASIC CANADA INC, Canada) and subsequently subcloned into the XhoI/NotI site of the MCS region of MFG-based retroviral vectors (FIG. 1C) in the manner described previously (Yang W et al, Int Immunol.2007; 19: 1083-93). All vector insertions encoding the TCR or TCR-CAR were verified by DNA sequencing.
Example 2: production of retroviruses encoding related target gene products
Phoenix amphoterics (293 cell-derived cell line with high calcium phosphate transfection efficiency) were transfected with 14 vectors, respectively (Table 1; FIG. 1, FIG. 2). According to standard procedures known in the art, such as those described in Yang W et al, Int immunol.2007; 19:1083-93 and Beaudoin EL et al J Virol Methods 2008; 148: 253-9, the virus supernatant (containing the viral particles) transfected with Phoenix cells was collected and used immediately to infect PG13 cells or other mouse cells, or stored at-80 ℃ for further use.
PG13 cells, a Virus Producer Cell (VPC) line, were infected with retroviral supernatants obtained from Phoenix cells, respectively, infected with the vector of interest. FACS-based cell sorting was performed to enrich for the corresponding vector-infected PG13 cells. Infected PG13 cells were enriched by cell sorting against human TCR V.beta.13.1 chain positive cells, or Nypep/A2 tetramer (tetramer/APC-HLA-A02: 01NY-ESO-1 (SLLMWITQC; SEQ ID NO:25) (Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA)) positive cells after staining with FITC anti-human TCR V.beta.13.1 antibody (EBIOSCIENCE, AFFYMETRIX and THERMO FISHER SCIENTIFIC), as described previously. High titer retroviral supernatants were produced using enriched VPC according to standard protocols well known in the art.
In one case, FACS analysis of infected PG13 cells was performed prior to FACS-based cell sorting enriched for TCR + or TCR-CAR + PG13 cells following viral infection. PG13 cells were infected with retroviral supernatants obtained from Phoenix cells infected with 3 vectors encoding NY-ESO-1TCR-CAR (NT22, NT24 and NT25), respectively. Infected PG13 cell line was analyzed by FACS for surface expression of tcrvb13.1 and binding of NYpep/a2 tetramer (CAR + cells were not previously enriched by FACS-based cell sorting). Live cells were gated for analysis. The intensity plots shown in FIG. 26 show positive staining of FITC anti-human TCRVb13.1 and APC-NYPep/A2 tetramer by single or double positive cells, respectively.
Example 3: generation of activated human T cells transduced with retrovirus alone
In Yang W et al, Int Immunol.2007; human T cell (ATC) -derived PBMCs activated with anti-CD 3 antibody were transduced with each of the following 7 vectors separately in the manner described in 1083-9: aNY-TCR (NT1, NT1b) and aNY-TCR-CAR (NT2, NT3, NT4 and NT5, NT6) (Table 1). 7 to 10 days after transduction, flow cytometry analysis WAs performed by combined staining with FITC-anti-human TCR V.beta.13.1 and tetramer/APC-HLA-A02: 01NY-ESO-1(SLLMWITQC, SEQ ID NO:5) (Fred Hutchinson Cancer Research Center, Seattle, WA). The results are shown in FIG. 27.
As shown in the figure, both the two second generation anti-NYESO-1/A2 TCR-CARs (NT2 and NT3) were expressed at significantly higher levels on transduced ATC (including CD4+ and CD8+ cells) cells, against both human tcrvb13.1 and NYpep/A2 tetramer-bound TCRs, compared to the native form (NT1 and NT1 b). Approximately 3.9% of untransduced cells (Untd) were also positive for TCRVb13.1 staining, but these cells were not significantly stained by the APC-NYPep/A2 tetramer.
Between 19.4-51.2% of activated T cells were found to bind to tetramer/APC-HLA-a 02:01 NY-ESO-1. The expression of TCR ν β 13.1 on its cell surface was measured based on FITC anti-human TCR ν β 13.1 antibody (EBIOSCIENCE/AFFYMETRIX, CA) staining and the similar percentage of activated T cells expressed based on correction of functional 1G 4a 95LY TCR domain on cell surface was measured based on positive tetramer staining with tetramer/PE-HLA-a 02:01NY-ESO-1 followed by FACS analysis. Further analysis showed that approximately half of the CAR + CD3+ cells of infected activated human T cells were CD4+ (52%) and CD8+ (48%).
Example 4: antigen-specific mediated cytokine IL-2 secretion using human ATC transduced against NY-ESO-1TCR or one of two TCR-based CARs
In this example, the activity of activated human T cells to induce cytokine secretion was examined by detecting the secretion of IL-2 in a medium in which T cells were co-cultured with HLA-A0201+ target cells in the absence and presence of various amounts of specific antigenic peptides.
Briefly, 0.5X10 in the absence or presence of various concentrations of the peptide NY-ESO-1(SLLMWITQC, SEQ ID NO:25, 10nM, 100nM and 2uM)5HLA-A0201+ target cells (T2 cells, 174x CEM. T2, ATCC CRL-1992)TM) Incubate in medium at 37 ℃ for 3 hours. Cells were washed twice to remove unbound peptide, then washed with 1.5x105The activated T cells were co-cultured for 24 hours in 200ul medium (complete RPMI 1640+ 10% FCS + Pen/Strep) (R-10) per well of a U-bottom 96-well plate, and transduced with the vectors NT1(NT1a), NT1b, NT2 and NT3, respectively (FIG. 2).
Culture supernatants were collected and IL-2 concentration was measured by ELISA detection kit (EBIOSCIENCES). The resulting IL-2 secretion data show that IL-2 is specifically secreted by activated T cells transduced with all three anti-NY-ESO-1 vectors, NT1(NT1a), NT1b, NT2 and NT3 (FIG. 2), respectively, after conjugation to NY-ESO-1 peptide pulsed T2 cells, but not T2 cells pulsed without NY-ESO-1 peptide or with negative control peptide.
It was found that T cells transduced with NT2 secreted the highest amount of IL-2 in these three, whereas T cells transduced with NT1 produced the lowest amount of IL-2 at the same peptide concentration of pulsed T2 cells. These results indicate that both TCR-CARs (NT2 and NT3) outperform NT1 in transduced TCR or TCR-CAR mediated T cell activation (fig. 2A).
Example 5: specific tumor cell killing using ATC transduced against NY ESO-1TCR or one of two TCR-based CARs
In this example, Saos-2 cells (A)HTB-85TM) Assays were performed to investigate the tumor cell killing activity of activated human T cells (ATC) transduced with anti-NY-ESO-1 TCR or TCR-based vectors NT1, NT1b, NT2 and NT3 (FIG. 2). The Saos-2 cell line (a human osteosarcoma cell line) is NY-ESO-1+ and HLA-A0201 +.
Briefly, 1x103Individual Saos-2 cells were pre-plated for 12 hours on 96-well flat-bottomed tissue culture plates. Then, 1x104Individual untransduced or transduced human T cells were added to each well. These T cells in complete RPMI 1640+ 10% FCS + Pen/Strep medium (R10) supplemented with 90IU rec hu IL-2/ml were either untransduced or have been transduced with the three vectors NT1, NT2 and NT3, respectively. Cells were cultured for up to 5 days (120hr) (FIG. 28). Surviving tumor target cells attached to the bottom of the wells, were much larger and distinct from T cells, and were observed and graded daily.
The killing activity of T cells was graded as: "+ ++", 75-100% kill; "+ ++", 75-100% kill; "+ ++", 50-75% kill; "+ +", 25-50% kill; "+", 0-25% kill was determined by comparing the number of surviving tumor cells in wells containing T cells with the number of surviving tumor cells in wells without T cells. At day 5 (120hr) after addition of T cells, photographs of representative views of wells were taken under a microscope. See fig. 28.
As shown in the figure, ATC (NT1(NT1a) and NT1b) expressing aNY-TCRa/b showed similar, highest tumor killing activity. ATC expressing NT2 showed slightly lower killing activity, while those with NT3 showed relatively lower killing activity. Untransduced ATC showed no significant killing effect. These results indicate that human T cells transduced with the four vectors, respectively, are capable of specifically killing Saos-2 tumor cells.
Example 6: antigen-specific mediated secretion of cytokines IL-2 and INF-gamma
In this example, an assay was performed to investigate cytokine secretion by ATC transduced with one of two anti-NY ESO-1 TCRs (NT1(NT1a) and NT1b) or one of two second generation TCR-based CARs (NT2 and NT 3).
ATCs transduced with NT1a, NT1b, NT2 or NT3 were prepared as described above. The percentage of NYpep/a2 positive T cells based on APC-NYpep/a2 tetramer staining by FACS (for NT1a, NT1b, NT2 and NT3) was adjusted to 20% using untransfected T cells ("undd") as described above. Then, 0.5x106Target cells (Saos-2 or control AspC1) or medium only (RPMI-1640+ 10% FCS + Pen/Strep) (R10) (without any IL-2 addition) with 2x10 transduced with NT1a, NT1b, NT2 or NT36One ATC was cultured in a total volume of 1.5mL of R10 (without any IL-2 addition) for 24 hours in a 24-well flat-bottomed tissue culture plate. The concentration of IL-2 or INF-gamma in the cell supernatants was measured by ELISA using a kit from BIOLEGENDS (Cat. Nos. 431804 and 430101). The results are shown in FIG. 29. As shown in FIG. 29A, T cells transduced with NT2 secreted the highest amounts of IL-2 (approximately 230pg/ml), while those using NT3 secreted moderate amounts of IL-2, those using each of the two native forms of anti-NY-ESO-1/A2 TCR (NT1 and NT1b) secreted the lowest amounts.
Example 7: percentage change of ATC NYPep/A2+ cells transduced with NT1, NT1b, NT2 and NT3
ATCs transduced with one of two anti-NY ESO-1 TCRs (NT1 and NT1b) or one of two second generation TCR-based CARs (NT2 and NT3) were prepared in the manner described above. The percentage of NYPep/A2 positive T cells based on APC-NYPep/A2 tetramer staining by FACS (for NT1a, NT1b, NT2 and NT3) was adjusted to 20% using undd T cells as described above.
On day 1, 1X106One target cell (Saos-2) and 4x106ATC transduced with NT1a, NT1b, NT2 or NT3 were co-cultured in a 12-well flat-bottomed tissue culture flask in a total volume of 4ml R10 containing 90IU IL-2/ml for 48 hours. On day 3, cells were transferred to a new 24-well plate. On day 6, cells from each well were split into two wells with R10 containing 180IU IL-2/ml in a total volume of 4 ml/well. On day 10, cells were harvested and analyzed by FACS for binding of the NYPep/A2 tetramer. The results are shown in FIG. 30. As shown in the figure, live cells were gated for analysis and the percentage of positive cells was shown. The percentage of NYpep/a2 positivity was found to increase most significantly in NT2 (about 32%), while a moderate increase was observed in NT3 (about 25%) and a decrease was observed in the NT1a and NT1b groups (about 17-19%).
Example 8: analysis of second and third Generation TCR-CAR
In this example, assays were performed on transduced activated human T cells to investigate the more native form of anti-NY-ESO-1 TCR and the second or third generation TCR-CAR.
First, the surface expression of TCR or TCR-CAR on transduced activated human T cells was investigated. Briefly, human T cells activated with anti-CD 3 antibody were either untransduced (Untd) or transduced with virus-containing supernatants from previously sorted anti-NY-ESO-1 TCR + (NT1 (also referred to as NT1a)) or anti-NY-ESO-1 TCR-CAR + ((NT 2; second generation), NT4 (vector 5; second generation), NT5 (vector 6; second generation) and NT24 (vector 11; third generation)) cell lines as described above. At 7 to 10 days post transduction, transduced T cells were analyzed by FACS for surface expression of TCRVb13.1 and binding of the NYPep/A2 tetramer. Live cells were gated for analysis. The results are shown in fig. 31A and 31B. As shown in the figure, T cells transduced with each of the three second generation anti-NY-ESO-1 TCR-CARs (NT2, NT4 and NT5) expressed the most and similar highest levels of tcrvb13.1 and NYpep/a2 tetramer-bound TCR or TCR-CAR as determined by FACS staining with tcrvb13.1 and NYpep/a2 tetramer. Those using the third generation TCR-CAR NT24 showed relatively low dual tcrvb13.1 and nypepp/a 2 tetramer binding + cells, while those using the native form TCR (NT1) showed the lowest percentage of dual tcrvb13.1 and nypepp/a 2 tetramer binding + cells.
Next, following conjugation to target cells, assays were performed to investigate the target cell killing ability of human ATC transduced with anti-NY ESO-1TCR or one of the four TCR-CAR. Briefly, ATCs transduced with NT1 (also known as NT1a), NT2, NT4, NT5 or NT24 were prepared as described above. Antigen-specific target cell killing against these activated human T cells was performed in an in vitro killing assay in which transduced human T cells (effectors) were co-cultured with Saos-2 tumor cells (targets) in the presence of the cytokine IL-2. In the experiments, human T cells transduced with NT1a, NT2, NT4, NT5 or NT24, respectively, were adjusted to 15% NYpep/a + using undd T cells, and then in the presence of cytokine IL-2 were adjusted to the following ratios 1:2 and 1:20 were co-cultured with Saos-2 tumor cells (target) for 12 hr. At the end of the incubation, T cell mediated cytotoxicity was measured using the Cayman 7-AAD/CFSE cell mediated cytotoxicity assay kit. The results are shown in figure 32, where the data are presented as mean ± s.d. from three samples in each group. As shown in figure 32, T cells transduced with one of NT1a and one of three TCR-based CARs (NT4, NT5 and NT24) comprising TCR signal 2 derived from 4-1BB showed the highest and similar target cell killing ability at these two E/T ratios (1:2 and 1: 20). Those using NT2 (second generation TCR-CRA, containing only TCR signal 2 derived from CD28) showed slightly lower killing ability. No significant target cell killing was seen with untransduced cells (Untd).
Again, cytokine secretion by human ATC transduced with TCR or TCR-based CARs after engagement with target cells was investigated. ATCs transduced with NT1a, NT2, NT4, NT5 or NT24 were prepared as described above. Transduced T cells were rescaled to 15% NYpep/a + using undd T cells. Then, 0.5x106One target Saos-2 cellOr medium alone (R10) with 2X106Transduced ATC or untransduced human T cells (Untd) were co-cultured in 24-well flat-bottomed tissue culture plates for 24 hours in a total volume of 1.5ml of R10 (without any IL-2 addition). The concentration of cytokines in the cell supernatants was measured by ELISA as described above. The results are shown in fig. 33A and 33B, with data expressed as mean ± s.d. from three samples in each group. As shown in fig. 33A, those using NT2 or NT24, both comprising 1(NT24) or 2(NT 2) TCR signaling 2 derived from human CD28, secreted the highest amounts of IL-2 (approximately 200pg/ml and 215pg/ml, respectively). Those using NT4 or NT5 (with TCR signal 2 from 4-1BB) produced moderate amounts (approximately 170pg/ml and 165pg/ml, respectively). Those that use the native form of anti-NY-ESO-1/A2 TCR (NT1) secrete minimal amounts. For IFN- γ, similar levels of IFN- γ were used with NT1 (also referred to as NT1a), NT2, NT4, NT5, and NT24 transduced ATC, respectively, after conjugation to the target Saos-2 cells, but not in the absence of target cells (R10 control), as shown in fig. 33B. Negative control cell Untd did not produce significant amounts of IL-2 or IFN-r in the presence or absence of target cells.
Finally, assays were performed to examine the change in the percentage and amplification of NYpep/a2+ ATC transduced with anti-NY ESO-1TCR or one of the four TCR-based CARs following conjugation to target cells. Briefly, ATC transduced with NT1a, NT2, NT4, NT5 or NT24 was adjusted to 15% NYpep/a + on day 1 using undd cells. Then, 1x106One target of Saos-2 cells with 6X106The ATC, which had been previously adjusted to 15% NYPep/A + cells and consisted of 0.9x10, was co-cultured in a total volume of 5ml of R10 containing 90IU IL-2/ml for 48 hours in a 12-well flat-bottomed tissue culture plate6NYPep/A + cells. At day 3, all cells were transferred to a new 12-well plate. On day 6 (6 days after the start of co-cultivation), the cells from each well were divided into two new wells with R10 containing 180IU IL-2/ml in a total volume of 5 ml/well. On day 10, suspension cells were collected, counted under microscopy using trypan blue staining, and analyzed by FACS for FITC-anti-human CD3 antibody and APC-NYPep/A2 tetramer. More than 95-98% of the viable cells were found to be CD3+ (data not shown). On live and CD3+ cellsGated for analysis. The results are shown in fig. 3A and 3B. The total Nypep/A2+ ATC is calculated according to the following formula:
the total number of viable cells as shown in FIG. 34A (by trypan blue staining) accounted for the% Npep/A2 + cells/Npep/A2 + ATC total.
As shown in figure 34A, the percentage of NYpep/a2 positive cells increased significantly from 15% to similar levels, ranging from 25.3% to 29.2%, in all four TCR-CARs (NT2, NT4, NT5, and NT 24); 28.4%, 27.1%, 25.3% and 29.2%, respectively. The increase in NT1a increased slightly to 19.1%. As shown in fig. 34B, the fold increases for NT2, NT4, NT5, and NT24 were 3.2, 3.1, 2.9, and 3.5, respectively, and 1.2 for NT 1.
Example 9: in vivo antitumor Activity
In this example, a xenograft mouse model was used to investigate the in vivo anti-tumor activity of T cells transduced with the vectors described herein.
Briefly, 5 days before adoptive cell transfer of effector T cells (day-5), in 100ul PBS (at e.g. 3x 10)6Individual cells/mouse), Saos-2 cells were injected subcutaneously into the posterior flank of SCID female mice. Activated or untransduced human T cells (Untd) transduced with retroviruses encoding NT1a, NT2, NT4 and NT24, respectively, were prepared as described above in fig. 31.
On day 0, the mice were divided into five groups of six mice each. ATC transduced with NT1a, NT2, NT4 or NT24 was adjusted to 15% NYpep/a + using untransduced T cells. Then, at 1x107At a dose of individual T cells/100 ul PBS/mouse, cells (containing CD4+ and CD8+ T cells) were administered intravenously via the tail vein.
The mice were monitored for tumor growth. Tumor size was measured using calipers and formula (tumor measurement (mm)3) Length x width x height x0.5236) to calculate tumor volume. Mice sacrificed due to oversized tumors or mice found to be dead were excluded from the collection and analysis of tumor size data. The results are shown in FIG. 35. As shown in the figure, all 6 mice (100%) of the NT 24-only group survived at day 54 after ACT administration. And, conversely, in0, 4 and 0 mice survived in NT1, NT2, NT4 and the undd groups, respectively. Furthermore, at the end of the experiment on day 62 after ATC, 3 of 6 mice in the NT24 group survived (50%), although none of 6 mice in the NT1, NT2, NT4 and unotd groups, respectively.
These in vivo data indicate that the third generation anti-NY-ESO-1/A2 TCR-CAR NT24 was significantly more potent than the second generation with TCR signal 2 from human CD28(NT2) alone or from 4-1BB (NT4) alone, and even more potent than native form NT1(NT1 a). The data also indicate that both of these second generation anti-NY-ESO-1 TCR-CAR show superior anti-tumor activity than native form NT1(NT1 a).
The foregoing examples and detailed description are intended to be illustrative, but not limiting, of the invention defined by the claims. As will be readily appreciated, numerous variations and combinations of the features set forth above may be utilized without departing from the present invention as set forth in the claims. Such variations are not to be regarded as a departure from the scope of the invention, and all such modifications are intended to be included within the scope of the following claims. All references cited herein are incorporated by reference in their entirety.
Sequence listing
<110> Yang, Wen
<120> compositions and methods for adoptive immunotherapy
<130> 180423.00200
<150> 62/877,331
<151> 2019-07-23
<150> 63/044,059
<151> 2020-06-25
<160> 73
<170> PatentIn version 3.5
<210> 1
<211> 275
<212> PRT
<213> Intelligent people
<400> 1
Met Glu Thr Leu Leu Gly Leu Leu Ile Leu Trp Leu Gln Leu Gln Trp
1 5 10 15
Val Ser Ser Lys Gln Glu Val Thr Gln Ile Pro Ala Ala Leu Ser Val
20 25 30
Pro Glu Gly Glu Asn Leu Val Leu Asn Cys Ser Phe Thr Asp Ser Ala
35 40 45
Ile Tyr Asn Leu Gln Trp Phe Arg Gln Asp Pro Gly Lys Gly Leu Thr
50 55 60
Ser Leu Leu Leu Ile Gln Ser Ser Gln Arg Glu Gln Thr Ser Gly Arg
65 70 75 80
Leu Asn Ala Ser Leu Asp Lys Ser Ser Gly Arg Ser Thr Leu Tyr Ile
85 90 95
Ala Ala Ser Gln Pro Gly Asp Ser Ala Thr Tyr Leu Cys Ala Val Arg
100 105 110
Pro Leu Tyr Gly Gly Ser Tyr Ile Pro Thr Phe Gly Arg Gly Thr Ser
115 120 125
Leu Ile Val His Pro Tyr Ile Gln Asn Pro Asp Pro Ala Val Tyr Gln
130 135 140
Leu Arg Asp Ser Lys Ser Ser Asp Lys Ser Val Cys Leu Phe Thr Asp
145 150 155 160
Phe Asp Ser Gln Thr Asn Val Ser Gln Ser Lys Asp Ser Asp Val Tyr
165 170 175
Ile Thr Asp Lys Thr Val Leu Asp Met Arg Ser Met Asp Phe Lys Ser
180 185 190
Asn Ser Ala Val Ala Trp Ser Asn Lys Ser Asp Phe Ala Cys Ala Asn
195 200 205
Ala Phe Asn Asn Ser Ile Ile Pro Glu Asp Thr Phe Phe Pro Ser Pro
210 215 220
Glu Ser Ser Cys Asp Val Lys Leu Val Glu Lys Ser Phe Glu Thr Asp
225 230 235 240
Thr Asn Leu Asn Phe Gln Asn Leu Ser Val Ile Gly Phe Arg Ile Leu
245 250 255
Leu Leu Lys Val Ala Gly Phe Asn Leu Leu Met Thr Leu Arg Leu Trp
260 265 270
Ser Ser Arg
275
<210> 2
<211> 311
<212> PRT
<213> Intelligent people
<400> 2
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Cys Gly Phe Thr Ser Glu Ser Tyr Gln Gln
260 265 270
Gly Val Leu Ser Ala Thr Ile Leu Tyr Glu Ile Leu Leu Gly Lys Ala
275 280 285
Thr Leu Tyr Ala Val Leu Val Ser Ala Leu Val Leu Met Ala Met Val
290 295 300
Lys Arg Lys Asp Ser Arg Gly
305 310
<210> 3
<211> 19
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 3
Ala Thr Asn Phe Ser Leu Leu Lys Gln Ala Gly Asp Val Glu Glu Asn
1 5 10 15
Pro Gly Pro
<210> 4
<211> 227
<212> PRT
<213> Intelligent people
<400> 4
Met Glu Thr Leu Leu Gly Leu Leu Ile Leu Trp Leu Gln Leu Gln Trp
1 5 10 15
Val Ser Ser Lys Gln Glu Val Thr Gln Ile Pro Ala Ala Leu Ser Val
20 25 30
Pro Glu Gly Glu Asn Leu Val Leu Asn Cys Ser Phe Thr Asp Ser Ala
35 40 45
Ile Tyr Asn Leu Gln Trp Phe Arg Gln Asp Pro Gly Lys Gly Leu Thr
50 55 60
Ser Leu Leu Leu Ile Gln Ser Ser Gln Arg Glu Gln Thr Ser Gly Arg
65 70 75 80
Leu Asn Ala Ser Leu Asp Lys Ser Ser Gly Arg Ser Thr Leu Tyr Ile
85 90 95
Ala Ala Ser Gln Pro Gly Asp Ser Ala Thr Tyr Leu Cys Ala Val Arg
100 105 110
Pro Leu Tyr Gly Gly Ser Tyr Ile Pro Thr Phe Gly Arg Gly Thr Ser
115 120 125
Leu Ile Val His Pro Tyr Ile Gln Asn Pro Asp Pro Ala Val Tyr Gln
130 135 140
Leu Arg Asp Ser Lys Ser Ser Asp Lys Ser Val Cys Leu Phe Thr Asp
145 150 155 160
Phe Asp Ser Gln Thr Asn Val Ser Gln Ser Lys Asp Ser Asp Val Tyr
165 170 175
Ile Thr Asp Lys Thr Val Leu Asp Met Arg Ser Met Asp Phe Lys Ser
180 185 190
Asn Ser Ala Val Ala Trp Ser Asn Lys Ser Asp Phe Ala Cys Ala Asn
195 200 205
Ala Phe Asn Asn Ser Ile Ile Pro Glu Asp Thr Phe Phe Pro Ser Pro
210 215 220
Glu Ser Ser
225
<210> 5
<211> 262
<212> PRT
<213> Intelligent people
<400> 5
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp
260
<210> 6
<211> 68
<212> PRT
<213> Intelligent people
<400> 6
Phe Trp Val Leu Val Val Val Gly Gly Val Leu Ala Cys Tyr Ser Leu
1 5 10 15
Leu Val Thr Val Ala Phe Ile Ile Phe Trp Val Arg Ser Lys Arg Ser
20 25 30
Arg Leu Leu His Ser Asp Tyr Met Asn Met Thr Pro Arg Arg Pro Gly
35 40 45
Pro Thr Arg Lys His Tyr Gln Pro Tyr Ala Pro Pro Arg Asp Phe Ala
50 55 60
Ala Tyr Arg Ser
65
<210> 7
<211> 112
<212> PRT
<213> Intelligent people
<400> 7
Arg Val Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln Gly
1 5 10 15
Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu Tyr
20 25 30
Asp Val Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met Gly Gly Lys
35 40 45
Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu Leu Gln Lys
50 55 60
Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys Gly Glu Arg
65 70 75 80
Arg Arg Gly Lys Gly His Asp Gly Leu Tyr Gln Gly Leu Ser Thr Ala
85 90 95
Thr Lys Asp Thr Tyr Asp Ala Leu His Met Gln Ala Leu Pro Pro Arg
100 105 110
<210> 8
<211> 55
<212> PRT
<213> Intelligent people
<400> 8
Lys Asn Arg Lys Ala Lys Ala Lys Pro Val Thr Arg Gly Ala Gly Ala
1 5 10 15
Gly Gly Arg Gln Arg Gly Gln Asn Lys Glu Arg Pro Pro Pro Val Pro
20 25 30
Asn Pro Asp Tyr Glu Pro Ile Arg Lys Gly Gln Arg Asp Leu Tyr Ser
35 40 45
Gly Leu Asn Gln Arg Arg Ile
50 55
<210> 9
<211> 612
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 9
Met Glu Thr Leu Leu Gly Leu Leu Ile Leu Trp Leu Gln Leu Gln Trp
1 5 10 15
Val Ser Ser Lys Gln Glu Val Thr Gln Ile Pro Ala Ala Leu Ser Val
20 25 30
Pro Glu Gly Glu Asn Leu Val Leu Asn Cys Ser Phe Thr Asp Ser Ala
35 40 45
Ile Tyr Asn Leu Gln Trp Phe Arg Gln Asp Pro Gly Lys Gly Leu Thr
50 55 60
Ser Leu Leu Leu Ile Gln Ser Ser Gln Arg Glu Gln Thr Ser Gly Arg
65 70 75 80
Leu Asn Ala Ser Leu Asp Lys Ser Ser Gly Arg Ser Thr Leu Tyr Ile
85 90 95
Ala Ala Ser Gln Pro Gly Asp Ser Ala Thr Tyr Leu Cys Ala Val Arg
100 105 110
Pro Leu Tyr Gly Gly Ser Tyr Ile Pro Thr Phe Gly Arg Gly Thr Ser
115 120 125
Leu Ile Val His Pro Tyr Ile Gln Asn Pro Asp Pro Ala Val Tyr Gln
130 135 140
Leu Arg Asp Ser Lys Ser Ser Asp Lys Ser Val Cys Leu Phe Thr Asp
145 150 155 160
Phe Asp Ser Gln Thr Asn Val Ser Gln Ser Lys Asp Ser Asp Val Tyr
165 170 175
Ile Thr Asp Lys Thr Val Leu Asp Met Arg Ser Met Asp Phe Lys Ser
180 185 190
Asn Ser Ala Val Ala Trp Ser Asn Lys Ser Asp Phe Ala Cys Ala Asn
195 200 205
Ala Phe Asn Asn Ser Ile Ile Pro Glu Asp Thr Phe Phe Pro Ser Pro
210 215 220
Glu Ser Ser Cys Asp Val Lys Leu Val Glu Lys Ser Phe Glu Thr Asp
225 230 235 240
Thr Asn Leu Asn Phe Gln Asn Leu Ser Val Ile Gly Phe Arg Ile Leu
245 250 255
Leu Leu Lys Val Ala Gly Phe Asn Leu Leu Met Thr Leu Arg Leu Trp
260 265 270
Ser Ser Arg Ala Lys Arg Ser Gly Ser Gly Ala Thr Asn Phe Ser Leu
275 280 285
Leu Lys Gln Ala Gly Asp Val Glu Glu Asn Pro Gly Pro Met Ser Ile
290 295 300
Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala Gly Pro Val
305 310 315 320
Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu Lys Thr Gly
325 330 335
Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His Glu Tyr Met
340 345 350
Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu Ile His Tyr
355 360 365
Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro Asn Gly Tyr
370 375 380
Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg Leu Leu Ser
385 390 395 400
Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser Ser Tyr Val
405 410 415
Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg Leu Thr Val
420 425 430
Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala Val Phe Glu
435 440 445
Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr Leu Val Cys
450 455 460
Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser Trp Trp Val
465 470 475 480
Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro Gln Pro Leu
485 490 495
Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu Ser Ser Arg
500 505 510
Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn His Phe Arg
515 520 525
Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu Trp Thr Gln
530 535 540
Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu Ala Trp Gly
545 550 555 560
Arg Ala Asp Cys Gly Phe Thr Ser Glu Ser Tyr Gln Gln Gly Val Leu
565 570 575
Ser Ala Thr Ile Leu Tyr Glu Ile Leu Leu Gly Lys Ala Thr Leu Tyr
580 585 590
Ala Val Leu Val Ser Ala Leu Val Leu Met Ala Met Val Lys Arg Lys
595 600 605
Asp Ser Arg Gly
610
<210> 10
<211> 883
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 10
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Gly Ser Pro Lys Phe Trp Val Leu Val Val
260 265 270
Val Gly Gly Val Leu Ala Cys Tyr Ser Leu Leu Val Thr Val Ala Phe
275 280 285
Ile Ile Phe Trp Val Arg Ser Lys Arg Ser Arg Leu Leu His Ser Asp
290 295 300
Tyr Met Asn Met Thr Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr
305 310 315 320
Gln Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala Tyr Arg Ser Arg Val
325 330 335
Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn
340 345 350
Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val
355 360 365
Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met Gly Gly Lys Pro Arg
370 375 380
Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu Leu Gln Lys Asp Lys
385 390 395 400
Met Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys Gly Glu Arg Arg Arg
405 410 415
Gly Lys Gly His Asp Gly Leu Tyr Gln Gly Leu Ser Thr Ala Thr Lys
420 425 430
Asp Thr Tyr Asp Ala Leu His Met Gln Ala Leu Pro Pro Arg Ala Lys
435 440 445
Arg Ser Gly Ser Gly Ala Thr Asn Phe Ser Leu Leu Lys Gln Ala Gly
450 455 460
Asp Val Glu Glu Asn Pro Gly Pro Met Glu Thr Leu Leu Gly Leu Leu
465 470 475 480
Ile Leu Trp Leu Gln Leu Gln Trp Val Ser Ser Lys Gln Glu Val Thr
485 490 495
Gln Ile Pro Ala Ala Leu Ser Val Pro Glu Gly Glu Asn Leu Val Leu
500 505 510
Asn Cys Ser Phe Thr Asp Ser Ala Ile Tyr Asn Leu Gln Trp Phe Arg
515 520 525
Gln Asp Pro Gly Lys Gly Leu Thr Ser Leu Leu Leu Ile Gln Ser Ser
530 535 540
Gln Arg Glu Gln Thr Ser Gly Arg Leu Asn Ala Ser Leu Asp Lys Ser
545 550 555 560
Ser Gly Arg Ser Thr Leu Tyr Ile Ala Ala Ser Gln Pro Gly Asp Ser
565 570 575
Ala Thr Tyr Leu Cys Ala Val Arg Pro Leu Tyr Gly Gly Ser Tyr Ile
580 585 590
Pro Thr Phe Gly Arg Gly Thr Ser Leu Ile Val His Pro Tyr Ile Gln
595 600 605
Asn Pro Asp Pro Ala Val Tyr Gln Leu Arg Asp Ser Lys Ser Ser Asp
610 615 620
Lys Ser Val Cys Leu Phe Thr Asp Phe Asp Ser Gln Thr Asn Val Ser
625 630 635 640
Gln Ser Lys Asp Ser Asp Val Tyr Ile Thr Asp Lys Thr Val Leu Asp
645 650 655
Met Arg Ser Met Asp Phe Lys Ser Asn Ser Ala Val Ala Trp Ser Asn
660 665 670
Lys Ser Asp Phe Ala Cys Ala Asn Ala Phe Asn Asn Ser Ile Ile Pro
675 680 685
Glu Asp Thr Phe Phe Pro Ser Pro Glu Ser Ser Gly Ser Pro Lys Phe
690 695 700
Trp Val Leu Val Val Val Gly Gly Val Leu Ala Cys Tyr Ser Leu Leu
705 710 715 720
Val Thr Val Ala Phe Ile Ile Phe Trp Val Arg Ser Lys Arg Ser Arg
725 730 735
Leu Leu His Ser Asp Tyr Met Asn Met Thr Pro Arg Arg Pro Gly Pro
740 745 750
Thr Arg Lys His Tyr Gln Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala
755 760 765
Tyr Arg Ser Arg Val Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr
770 775 780
Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg
785 790 795 800
Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met
805 810 815
Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu
820 825 830
Leu Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys
835 840 845
Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu Tyr Gln Gly Leu
850 855 860
Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His Met Gln Ala Leu
865 870 875 880
Pro Pro Arg
<210> 11
<211> 769
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 11
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Gly Ser Pro Lys Phe Trp Val Leu Val Val
260 265 270
Val Gly Gly Val Leu Ala Cys Tyr Ser Leu Leu Val Thr Val Ala Phe
275 280 285
Ile Ile Phe Trp Val Arg Ser Lys Arg Ser Arg Leu Leu His Ser Asp
290 295 300
Tyr Met Asn Met Thr Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr
305 310 315 320
Gln Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala Tyr Arg Ser Lys Asn
325 330 335
Arg Lys Ala Lys Ala Lys Pro Val Thr Arg Gly Ala Gly Ala Gly Gly
340 345 350
Arg Gln Arg Gly Gln Asn Lys Glu Arg Pro Pro Pro Val Pro Asn Pro
355 360 365
Asp Tyr Glu Pro Ile Arg Lys Gly Gln Arg Asp Leu Tyr Ser Gly Leu
370 375 380
Asn Gln Arg Arg Ile Ala Lys Arg Ser Gly Ser Gly Ala Thr Asn Phe
385 390 395 400
Ser Leu Leu Lys Gln Ala Gly Asp Val Glu Glu Asn Pro Gly Pro Met
405 410 415
Glu Thr Leu Leu Gly Leu Leu Ile Leu Trp Leu Gln Leu Gln Trp Val
420 425 430
Ser Ser Lys Gln Glu Val Thr Gln Ile Pro Ala Ala Leu Ser Val Pro
435 440 445
Glu Gly Glu Asn Leu Val Leu Asn Cys Ser Phe Thr Asp Ser Ala Ile
450 455 460
Tyr Asn Leu Gln Trp Phe Arg Gln Asp Pro Gly Lys Gly Leu Thr Ser
465 470 475 480
Leu Leu Leu Ile Gln Ser Ser Gln Arg Glu Gln Thr Ser Gly Arg Leu
485 490 495
Asn Ala Ser Leu Asp Lys Ser Ser Gly Arg Ser Thr Leu Tyr Ile Ala
500 505 510
Ala Ser Gln Pro Gly Asp Ser Ala Thr Tyr Leu Cys Ala Val Arg Pro
515 520 525
Leu Tyr Gly Gly Ser Tyr Ile Pro Thr Phe Gly Arg Gly Thr Ser Leu
530 535 540
Ile Val His Pro Tyr Ile Gln Asn Pro Asp Pro Ala Val Tyr Gln Leu
545 550 555 560
Arg Asp Ser Lys Ser Ser Asp Lys Ser Val Cys Leu Phe Thr Asp Phe
565 570 575
Asp Ser Gln Thr Asn Val Ser Gln Ser Lys Asp Ser Asp Val Tyr Ile
580 585 590
Thr Asp Lys Thr Val Leu Asp Met Arg Ser Met Asp Phe Lys Ser Asn
595 600 605
Ser Ala Val Ala Trp Ser Asn Lys Ser Asp Phe Ala Cys Ala Asn Ala
610 615 620
Phe Asn Asn Ser Ile Ile Pro Glu Asp Thr Phe Phe Pro Ser Pro Glu
625 630 635 640
Ser Ser Gly Ser Pro Lys Phe Trp Val Leu Val Val Val Gly Gly Val
645 650 655
Leu Ala Cys Tyr Ser Leu Leu Val Thr Val Ala Phe Ile Ile Phe Trp
660 665 670
Val Arg Ser Lys Arg Ser Arg Leu Leu His Ser Asp Tyr Met Asn Met
675 680 685
Thr Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr Gln Pro Tyr Ala
690 695 700
Pro Pro Arg Asp Phe Ala Ala Tyr Arg Ser Lys Asn Arg Lys Ala Lys
705 710 715 720
Ala Lys Pro Val Thr Arg Gly Ala Gly Ala Gly Gly Arg Gln Arg Gly
725 730 735
Gln Asn Lys Glu Arg Pro Pro Pro Val Pro Asn Pro Asp Tyr Glu Pro
740 745 750
Ile Arg Lys Gly Gln Arg Asp Leu Tyr Ser Gly Leu Asn Gln Arg Arg
755 760 765
Ile
<210> 12
<211> 1839
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 12
atggagaccc tcttgggcct gcttatcctt tggctgcagc tgcaatgggt gagcagcaaa 60
caggaggtga cgcagattcc tgcagctctg agtgtcccag aaggagaaaa cttggttctc 120
aactgcagtt tcactgatag cgctatttac aacctccagt ggtttaggca ggaccctggg 180
aaaggtctca catctctgtt gcttattcag tcaagtcaga gagagcaaac aagtggaaga 240
cttaatgcct cgctggataa atcatcagga cgtagtactt tatacattgc agcttctcag 300
cctggtgact cagccaccta cctctgtgct gtgaggcccc tgtacggagg aagctacata 360
cctacatttg gaagaggaac cagccttatt gttcatccgt atatccagaa ccctgaccct 420
gccgtgtacc agctgagaga ctctaaatcc agtgacaagt ctgtctgcct attcaccgat 480
tttgattctc aaacaaatgt gtcacaaagt aaggattctg atgtgtatat cacagacaaa 540
actgtgctag acatgaggtc tatggacttc aagagcaaca gtgctgtggc ctggagcaac 600
aaatctgact ttgcatgtgc aaacgccttc aacaacagca ttattccaga agacaccttc 660
ttccccagcc cagaaagttc ctgtgatgtc aagctggtcg agaaaagctt tgaaacagat 720
acgaacctaa actttcaaaa cctgtcagtg attgggttcc gaatcctcct cctgaaagtg 780
gccgggttta atctgctcat gacgctgcgg ctgtggtcca gccgggccaa gcggtctggg 840
tctggggcca ccaacttcag cctgctgaag caggccggcg acgtggagga gaaccccggc 900
cccatgagca tcggcctcct gtgctgtgca gccttgtctc tcctgtgggc aggtccagtg 960
aatgctggtg tcactcagac cccaaaattc caggtcctga agacaggaca gagcatgaca 1020
ctgcagtgtg cccaggatat gaaccatgaa tacatgtcct ggtatcgaca agacccaggc 1080
atggggctga ggctgattca ttactcagtt ggtgctggta tcactgacca aggagaagtc 1140
cccaatggct acaatgtctc cagatcaacc acagaggatt tcccgctcag gctgctgtcg 1200
gctgctccct cccagacatc tgtgtacttc tgtgccagca gttacgtcgg gaacaccggg 1260
gagctgtttt ttggagaagg ctctaggctg accgtactgg aggacctgaa aaacgtgttc 1320
ccacccgagg tcgctgtgtt tgagccatca gaagcagaga tctcccacac ccaaaaggcc 1380
acactggtgt gcctggccac aggcttctac cccgaccacg tggagctgag ctggtgggtg 1440
aatgggaagg aggtgcacag tggggtcagc acagacccgc agcccctcaa ggagcagccc 1500
gccctcaatg actccagata cgctctgagc agccgcctga gggtctcggc caccttctgg 1560
caggaccccc gcaaccactt ccgctgtcaa gtccagttct acgggctctc ggagaatgac 1620
gagtggaccc aggatagggc caaacccgtc acccagatcg tcagcgccga ggcctggggt 1680
agagcagact gtggcttcac ctccgagtct taccagcaag gggtcctgtc tgccaccatc 1740
ctctatgaga tcttgctagg gaaggccacc ttgtatgccg tgctggtcag tgccctcgtg 1800
ctgatggcta tggtcaagag aaaggattcc agaggctaa 1839
<210> 13
<211> 2652
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 13
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagacggct ctcctaagtt ctgggttctc gtcgtcgtgg gaggtgtgtt agcatgttac 840
tctctcttgg ttactgtcgc tttcataatc ttttgggtcc gctcaaaacg ctctcgcttg 900
ttacattccg attatatgaa tatgacacct aggagacctg gcccgactag gaaacactat 960
caaccttacg cacctcccag agattttgct gcttacagga gtcgggtcaa attttcacgc 1020
tccgctgatg ctcctgccta tcaacaaggg caaaatcaat tgtacaatga attgaacttg 1080
ggtagaaggg aagaatatga cgtgctcgat aaacggaggg ggagagatcc agaaatgggc 1140
ggtaaaccac ggcgcaaaaa tccacaagag ggattgtata acgagctcca aaaggacaaa 1200
atggcagaag cttattcaga aataggaatg aagggggaaa ggagacgagg taaaggtcat 1260
gacggattgt atcaaggatt gtcaaccgct actaaagata catatgatgc tttgcatatg 1320
caagctttgc ctcccagagc caagcggtct gggtctgggg ccaccaactt cagcctgctg 1380
aagcaggccg gcgacgtgga ggagaacccc ggccccatgg agaccctctt gggcctgctt 1440
atcctttggc tgcagctgca atgggtgagc agcaaacagg aggtgacgca gattcctgca 1500
gctctgagtg tcccagaagg agaaaacttg gttctcaact gcagtttcac tgatagcgct 1560
atttacaacc tccagtggtt taggcaggac cctgggaaag gtctcacatc tctgttgctt 1620
attcagtcaa gtcagagaga gcaaacaagt ggaagactta atgcctcgct ggataaatca 1680
tcaggacgta gtactttata cattgcagct tctcagcctg gtgactcagc cacctacctc 1740
tgtgctgtga ggcccctgta cggaggaagc tacataccta catttggaag aggaaccagc 1800
cttattgttc atccgtatat ccagaaccct gaccctgccg tgtaccagct gagagactct 1860
aaatccagtg acaagtctgt ctgcctattc accgattttg attctcaaac aaatgtgtca 1920
caaagtaagg attctgatgt gtatatcaca gacaaaactg tgctagacat gaggtctatg 1980
gacttcaaga gcaacagtgc tgtggcctgg agcaacaaat ctgactttgc atgtgcaaac 2040
gccttcaaca acagcattat tccagaagac accttcttcc ccagcccaga aagttccggc 2100
tccccaaaat tttgggtgct ggtggtggtt ggtggagtcc tggcttgcta tagcttgcta 2160
gtaacagtgg cctttattat tttctgggtg aggagtaaga ggagcaggct cctgcacagt 2220
gactacatga acatgactcc ccgccgcccc gggcccaccc gcaagcatta ccagccctat 2280
gccccaccac gcgacttcgc agcctatcgc tccagagtga agttcagcag gagcgcagac 2340
gcccccgcgt accagcaggg ccagaaccag ctctataacg agctcaatct aggacgaaga 2400
gaggagtacg atgttttgga caagagacgt ggccgggacc ctgagatggg gggaaagccg 2460
agaaggaaga accctcagga aggcctgtac aatgaactgc agaaagataa gatggcggag 2520
gcctacagtg agattgggat gaaaggcgag cgccggaggg gcaaggggca cgatggcctt 2580
taccagggtc tcagtacagc caccaaggac acctacgacg cccttcacat gcaggccctg 2640
ccccctcgct aa 2652
<210> 14
<211> 2310
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 14
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagacggct ctcctaagtt ctgggttctc gtcgtcgtgg gaggtgtgtt agcatgttac 840
tctctcttgg ttactgtcgc tttcataatc ttttgggtcc gctcaaaacg ctctcgcttg 900
ttacattccg attatatgaa tatgacacct aggagacctg gcccgactag gaaacactat 960
caaccttacg cacctcccag agattttgct gcttacagga gtaaaaaccg caaagctaaa 1020
gctaaacccg tcactagggg ggccggagca ggagggcgcc agcgcggtca gaataaagaa 1080
cgccctcctc ccgtccctaa tcctgattac gaaccgatta gaaaggggca aagagatctc 1140
tacagcggac tcaaccaacg gagaattgcc aagcggtctg ggtctggggc caccaacttc 1200
agcctgctga agcaggccgg cgacgtggag gagaaccccg gccccatgga gaccctcttg 1260
ggcctgctta tcctttggct gcagctgcaa tgggtgagca gcaaacagga ggtgacgcag 1320
attcctgcag ctctgagtgt cccagaagga gaaaacttgg ttctcaactg cagtttcact 1380
gatagcgcta tttacaacct ccagtggttt aggcaggacc ctgggaaagg tctcacatct 1440
ctgttgctta ttcagtcaag tcagagagag caaacaagtg gaagacttaa tgcctcgctg 1500
gataaatcat caggacgtag tactttatac attgcagctt ctcagcctgg tgactcagcc 1560
acctacctct gtgctgtgag gcccctgtac ggaggaagct acatacctac atttggaaga 1620
ggaaccagcc ttattgttca tccgtatatc cagaaccctg accctgccgt gtaccagctg 1680
agagactcta aatccagtga caagtctgtc tgcctattca ccgattttga ttctcaaaca 1740
aatgtgtcac aaagtaagga ttctgatgtg tatatcacag acaaaactgt gctagacatg 1800
aggtctatgg acttcaagag caacagtgct gtggcctgga gcaacaaatc tgactttgca 1860
tgtgcaaacg ccttcaacaa cagcattatt ccagaagaca ccttcttccc cagcccagaa 1920
agttccggct ccccaaaatt ttgggtgctg gtggtggttg gtggagtcct ggcttgctat 1980
agcttgctag taacagtggc ctttattatt ttctgggtga ggagtaagag gagcaggctc 2040
ctgcacagtg actacatgaa catgactccc cgccgccccg ggcccacccg caagcattac 2100
cagccctatg ccccaccacg cgacttcgca gcctatcgct ccaagaatag aaaggccaag 2160
gccaagcctg tgacacgagg agcgggtgct ggcggcaggc aaaggggaca aaacaaggag 2220
aggccaccac ctgttcccaa cccagactat gagcccatcc gcaaaggcca gcgggacctg 2280
tattctggcc tgaatcagag acgcatctaa 2310
<210> 15
<211> 141
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 15
Met Asn Met Thr Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr Gln
1 5 10 15
Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala Tyr Arg Ser Arg Val Lys
20 25 30
Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn Gln
35 40 45
Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val Leu
50 55 60
Asp Lys Arg Arg Gly Arg Asp Pro Glu Met Gly Gly Lys Pro Arg Arg
65 70 75 80
Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu Leu Gln Lys Asp Lys Met
85 90 95
Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys Gly Glu Arg Arg Arg Gly
100 105 110
Lys Gly His Asp Gly Leu Tyr Gln Gly Leu Ser Thr Ala Thr Lys Asp
115 120 125
Thr Tyr Asp Ala Leu His Met Gln Ala Leu Pro Pro Arg
130 135 140
<210> 16
<211> 540
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 16
ttttgggtgc tggtggtggt tggtggagtc ctggcttgct atagcttgct agtaacagtg 60
gcctttatta ttttctgggt gaggagtaag aggagcaggc tcctgcacag tgactacatg 120
aacatgactc cccgccgccc cgggcccacc cgcaagcatt accagcccta tgccccacca 180
cgcgacttcg cagcctatcg ctccagagtg aagttcagca ggagcgcaga cgcccccgcg 240
taccagcagg gccagaacca gctctataac gagctcaatc taggacgaag agaggagtac 300
gatgttttgg acaagagacg tggccgggac cctgagatgg ggggaaagcc gagaaggaag 360
aaccctcagg aaggcctgta caatgaactg cagaaagata agatggcgga ggcctacagt 420
gagattggga tgaaaggcga gcgccggagg ggcaaggggc acgatggcct ttaccagggt 480
ctcagtacag ccaccaagga cacctacgac gcccttcaca tgcaggccct gccccctcgc 540
<210> 17
<211> 204
<212> DNA
<213> Intelligent people
<400> 17
ttttgggtgc tggtggtggt tggtggagtc ctggcttgct atagcttgct agtaacagtg 60
gcctttatta ttttctgggt gaggagtaag aggagcaggc tcctgcacag tgactacatg 120
aacatgactc cccgccgccc cgggcccacc cgcaagcatt accagcccta tgccccacca 180
cgcgacttcg cagcctatcg ctcc 204
<210> 18
<211> 204
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 18
ttctgggttc tcgtcgtcgt gggaggtgtg ttagcatgtt actctctctt ggttactgtc 60
gctttcataa tcttttgggt ccgctcaaaa cgctctcgct tgttacattc cgattatatg 120
aatatgacac ctaggagacc tggcccgact aggaaacact atcaacctta cgcacctccc 180
agagattttg ctgcttacag gagt 204
<210> 19
<211> 336
<212> DNA
<213> Intelligent people
<400> 19
agagtgaagt tcagcaggag cgcagacgcc cccgcgtacc agcagggcca gaaccagctc 60
tataacgagc tcaatctagg acgaagagag gagtacgatg ttttggacaa gagacgtggc 120
cgggaccctg agatgggggg aaagccgaga aggaagaacc ctcaggaagg cctgtacaat 180
gaactgcaga aagataagat ggcggaggcc tacagtgaga ttgggatgaa aggcgagcgc 240
cggaggggca aggggcacga tggcctttac cagggtctca gtacagccac caaggacacc 300
tacgacgccc ttcacatgca ggccctgccc cctcgc 336
<210> 20
<211> 336
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 20
cgggtcaaat tttcacgctc cgctgatgct cctgcctatc aacaagggca aaatcaattg 60
tacaatgaat tgaacttggg tagaagggaa gaatatgacg tgctcgataa acggaggggg 120
agagatccag aaatgggcgg taaaccacgg cgcaaaaatc cacaagaggg attgtataac 180
gagctccaaa aggacaaaat ggcagaagct tattcagaaa taggaatgaa gggggaaagg 240
agacgaggta aaggtcatga cggattgtat caaggattgt caaccgctac taaagataca 300
tatgatgctt tgcatatgca agctttgcct cccaga 336
<210> 21
<211> 540
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 21
ttctgggttc tcgtcgtcgt gggaggtgtg ttagcatgtt actctctctt ggttactgtc 60
gctttcataa tcttttgggt ccgctcaaaa cgctctcgct tgttacattc cgattatatg 120
aatatgacac ctaggagacc tggcccgact aggaaacact atcaacctta cgcacctccc 180
agagattttg ctgcttacag gagtcgggtc aaattttcac gctccgctga tgctcctgcc 240
tatcaacaag ggcaaaatca attgtacaat gaattgaact tgggtagaag ggaagaatat 300
gacgtgctcg ataaacggag ggggagagat ccagaaatgg gcggtaaacc acggcgcaaa 360
aatccacaag agggattgta taacgagctc caaaaggaca aaatggcaga agcttattca 420
gaaataggaa tgaaggggga aaggagacga ggtaaaggtc atgacggatt gtatcaagga 480
ttgtcaaccg ctactaaaga tacatatgat gctttgcata tgcaagcttt gcctcccaga 540
<210> 22
<211> 165
<212> DNA
<213> Intelligent people
<400> 22
aagaatagaa aggccaaggc caagcctgtg acacgaggag cgggtgctgg cggcaggcaa 60
aggggacaaa acaaggagag gccaccacct gttcccaacc cagactatga gcccatccgg 120
aaaggccagc gggacctgta ttctggcctg aatcagagac gcatc 165
<210> 23
<211> 165
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 23
aagaatagaa aggccaaggc caagcctgtg acacgaggag cgggtgctgg cggcaggcaa 60
aggggacaaa acaaggagag gccaccacct gttcccaacc cagactatga gcccatccgc 120
aaaggccagc gggacctgta ttctggcctg aatcagagac gcatc 165
<210> 24
<211> 165
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 24
aaaaaccgca aagctaaagc taaacccgtc actagggggg ccggagcagg agggcgccag 60
cgcggtcaga ataaagaacg ccctcctccc gtccctaatc ctgattacga accgattaga 120
aaggggcaaa gagatctcta cagcggactc aaccaacgga gaatt 165
<210> 25
<211> 9
<212> PRT
<213> Intelligent people
<400> 25
Ser Leu Leu Met Trp Ile Thr Gln Cys
1 5
<210> 26
<211> 40
<212> PRT
<213> Intelligent people
<400> 26
Lys Ile Glu Val Met Tyr Pro Pro Pro Tyr Leu Asp Asn Glu Lys Ser
1 5 10 15
Asn Gly Thr Ile Ile His Val Lys Gly Lys His Leu Cys Pro Ser Pro
20 25 30
Leu Phe Pro Gly Pro Ser Lys Pro
35 40
<210> 27
<211> 27
<212> PRT
<213> Intelligent people
<400> 27
Phe Trp Val Leu Val Val Val Gly Gly Val Leu Ala Cys Tyr Ser Leu
1 5 10 15
Leu Val Thr Val Ala Phe Ile Ile Phe Trp Val
20 25
<210> 28
<211> 41
<212> PRT
<213> Intelligent people
<400> 28
Arg Ser Lys Arg Ser Arg Leu Leu His Ser Asp Tyr Met Asn Met Thr
1 5 10 15
Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr Gln Pro Tyr Ala Pro
20 25 30
Pro Arg Asp Phe Ala Ala Tyr Arg Ser
35 40
<210> 29
<211> 42
<212> PRT
<213> Intelligent people
<400> 29
Lys Arg Gly Arg Lys Lys Leu Leu Tyr Ile Phe Lys Gln Pro Phe Met
1 5 10 15
Arg Pro Val Gln Thr Thr Gln Glu Glu Asp Gly Cys Ser Cys Arg Phe
20 25 30
Pro Glu Glu Glu Glu Gly Gly Cys Glu Leu
35 40
<210> 30
<211> 46
<212> PRT
<213> Intelligent people
<400> 30
Ala Lys Pro Thr Thr Thr Pro Ala Pro Arg Pro Pro Thr Pro Ala Pro
1 5 10 15
Thr Ile Ala Ser Gln Pro Leu Ser Leu Arg Pro Glu Ala Cys Arg Pro
20 25 30
Ala Ala Gly Gly Ala Val His Thr Arg Gly Leu Asp Phe Ala
35 40 45
<210> 31
<211> 26
<212> PRT
<213> Intelligent people
<400> 31
Cys Asp Ile Tyr Ile Trp Ala Pro Leu Ala Gly Thr Cys Gly Val Leu
1 5 10 15
Leu Leu Ser Leu Val Ile Thr Leu Tyr Cys
20 25
<210> 32
<211> 883
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 32
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Gly Ser Pro Lys Cys Asp Ile Tyr Ile Trp
260 265 270
Ala Pro Leu Ala Gly Thr Cys Gly Val Leu Leu Leu Ser Leu Val Ile
275 280 285
Thr Leu Tyr Cys Lys Arg Gly Arg Lys Lys Leu Leu Tyr Ile Phe Lys
290 295 300
Gln Pro Phe Met Arg Pro Val Gln Thr Thr Gln Glu Glu Asp Gly Cys
305 310 315 320
Ser Cys Arg Phe Pro Glu Glu Glu Glu Gly Gly Cys Glu Leu Arg Val
325 330 335
Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn
340 345 350
Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val
355 360 365
Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met Gly Gly Lys Pro Arg
370 375 380
Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu Leu Gln Lys Asp Lys
385 390 395 400
Met Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys Gly Glu Arg Arg Arg
405 410 415
Gly Lys Gly His Asp Gly Leu Tyr Gln Gly Leu Ser Thr Ala Thr Lys
420 425 430
Asp Thr Tyr Asp Ala Leu His Met Gln Ala Leu Pro Pro Arg Ala Lys
435 440 445
Arg Ser Gly Ser Gly Ala Thr Asn Phe Ser Leu Leu Lys Gln Ala Gly
450 455 460
Asp Val Glu Glu Asn Pro Gly Pro Met Glu Thr Leu Leu Gly Leu Leu
465 470 475 480
Ile Leu Trp Leu Gln Leu Gln Trp Val Ser Ser Lys Gln Glu Val Thr
485 490 495
Gln Ile Pro Ala Ala Leu Ser Val Pro Glu Gly Glu Asn Leu Val Leu
500 505 510
Asn Cys Ser Phe Thr Asp Ser Ala Ile Tyr Asn Leu Gln Trp Phe Arg
515 520 525
Gln Asp Pro Gly Lys Gly Leu Thr Ser Leu Leu Leu Ile Gln Ser Ser
530 535 540
Gln Arg Glu Gln Thr Ser Gly Arg Leu Asn Ala Ser Leu Asp Lys Ser
545 550 555 560
Ser Gly Arg Ser Thr Leu Tyr Ile Ala Ala Ser Gln Pro Gly Asp Ser
565 570 575
Ala Thr Tyr Leu Cys Ala Val Arg Pro Leu Tyr Gly Gly Ser Tyr Ile
580 585 590
Pro Thr Phe Gly Arg Gly Thr Ser Leu Ile Val His Pro Tyr Ile Gln
595 600 605
Asn Pro Asp Pro Ala Val Tyr Gln Leu Arg Asp Ser Lys Ser Ser Asp
610 615 620
Lys Ser Val Cys Leu Phe Thr Asp Phe Asp Ser Gln Thr Asn Val Ser
625 630 635 640
Gln Ser Lys Asp Ser Asp Val Tyr Ile Thr Asp Lys Thr Val Leu Asp
645 650 655
Met Arg Ser Met Asp Phe Lys Ser Asn Ser Ala Val Ala Trp Ser Asn
660 665 670
Lys Ser Asp Phe Ala Cys Ala Asn Ala Phe Asn Asn Ser Ile Ile Pro
675 680 685
Glu Asp Thr Phe Phe Pro Ser Pro Glu Ser Ser Gly Ser Pro Lys Cys
690 695 700
Asp Ile Tyr Ile Trp Ala Pro Leu Ala Gly Thr Cys Gly Val Leu Leu
705 710 715 720
Leu Ser Leu Val Ile Thr Leu Tyr Cys Lys Arg Gly Arg Lys Lys Leu
725 730 735
Leu Tyr Ile Phe Lys Gln Pro Phe Met Arg Pro Val Gln Thr Thr Gln
740 745 750
Glu Glu Asp Gly Cys Ser Cys Arg Phe Pro Glu Glu Glu Glu Gly Gly
755 760 765
Cys Glu Leu Arg Val Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr
770 775 780
Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg
785 790 795 800
Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met
805 810 815
Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu
820 825 830
Leu Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys
835 840 845
Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu Tyr Gln Gly Leu
850 855 860
Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His Met Gln Ala Leu
865 870 875 880
Pro Pro Arg
<210> 33
<211> 885
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 33
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Gly Ser Pro Lys Phe Trp Val Leu Val Val
260 265 270
Val Gly Gly Val Leu Ala Cys Tyr Ser Leu Leu Val Thr Val Ala Phe
275 280 285
Ile Ile Phe Trp Val Lys Arg Gly Arg Lys Lys Leu Leu Tyr Ile Phe
290 295 300
Lys Gln Pro Phe Met Arg Pro Val Gln Thr Thr Gln Glu Glu Asp Gly
305 310 315 320
Cys Ser Cys Arg Phe Pro Glu Glu Glu Glu Gly Gly Cys Glu Leu Arg
325 330 335
Val Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln Gly Gln
340 345 350
Asn Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu Tyr Asp
355 360 365
Val Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met Gly Gly Lys Pro
370 375 380
Arg Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu Leu Gln Lys Asp
385 390 395 400
Lys Met Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys Gly Glu Arg Arg
405 410 415
Arg Gly Lys Gly His Asp Gly Leu Tyr Gln Gly Leu Ser Thr Ala Thr
420 425 430
Lys Asp Thr Tyr Asp Ala Leu His Met Gln Ala Leu Pro Pro Arg Ala
435 440 445
Lys Arg Ser Gly Ser Gly Ala Thr Asn Phe Ser Leu Leu Lys Gln Ala
450 455 460
Gly Asp Val Glu Glu Asn Pro Gly Pro Met Glu Thr Leu Leu Gly Leu
465 470 475 480
Leu Ile Leu Trp Leu Gln Leu Gln Trp Val Ser Ser Lys Gln Glu Val
485 490 495
Thr Gln Ile Pro Ala Ala Leu Ser Val Pro Glu Gly Glu Asn Leu Val
500 505 510
Leu Asn Cys Ser Phe Thr Asp Ser Ala Ile Tyr Asn Leu Gln Trp Phe
515 520 525
Arg Gln Asp Pro Gly Lys Gly Leu Thr Ser Leu Leu Leu Ile Gln Ser
530 535 540
Ser Gln Arg Glu Gln Thr Ser Gly Arg Leu Asn Ala Ser Leu Asp Lys
545 550 555 560
Ser Ser Gly Arg Ser Thr Leu Tyr Ile Ala Ala Ser Gln Pro Gly Asp
565 570 575
Ser Ala Thr Tyr Leu Cys Ala Val Arg Pro Leu Tyr Gly Gly Ser Tyr
580 585 590
Ile Pro Thr Phe Gly Arg Gly Thr Ser Leu Ile Val His Pro Tyr Ile
595 600 605
Gln Asn Pro Asp Pro Ala Val Tyr Gln Leu Arg Asp Ser Lys Ser Ser
610 615 620
Asp Lys Ser Val Cys Leu Phe Thr Asp Phe Asp Ser Gln Thr Asn Val
625 630 635 640
Ser Gln Ser Lys Asp Ser Asp Val Tyr Ile Thr Asp Lys Thr Val Leu
645 650 655
Asp Met Arg Ser Met Asp Phe Lys Ser Asn Ser Ala Val Ala Trp Ser
660 665 670
Asn Lys Ser Asp Phe Ala Cys Ala Asn Ala Phe Asn Asn Ser Ile Ile
675 680 685
Pro Glu Asp Thr Phe Phe Pro Ser Pro Glu Ser Ser Gly Ser Pro Lys
690 695 700
Phe Trp Val Leu Val Val Val Gly Gly Val Leu Ala Cys Tyr Ser Leu
705 710 715 720
Leu Val Thr Val Ala Phe Ile Ile Phe Trp Val Lys Arg Gly Arg Lys
725 730 735
Lys Leu Leu Tyr Ile Phe Lys Gln Pro Phe Met Arg Pro Val Gln Thr
740 745 750
Thr Gln Glu Glu Asp Gly Cys Ser Cys Arg Phe Pro Glu Glu Glu Glu
755 760 765
Gly Gly Cys Glu Leu Arg Val Lys Phe Ser Arg Ser Ala Asp Ala Pro
770 775 780
Ala Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu Gly
785 790 795 800
Arg Arg Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg Asp Pro
805 810 815
Glu Met Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu Tyr
820 825 830
Asn Glu Leu Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile Gly
835 840 845
Met Lys Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu Tyr Gln
850 855 860
Gly Leu Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His Met Gln
865 870 875 880
Ala Leu Pro Pro Arg
885
<210> 34
<211> 967
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 34
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Gly Ser Pro Lys Phe Trp Val Leu Val Val
260 265 270
Val Gly Gly Val Leu Ala Cys Tyr Ser Leu Leu Val Thr Val Ala Phe
275 280 285
Ile Ile Phe Trp Val Arg Ser Lys Arg Ser Arg Leu Leu His Ser Asp
290 295 300
Tyr Met Asn Met Thr Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr
305 310 315 320
Gln Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala Tyr Arg Ser Lys Arg
325 330 335
Gly Arg Lys Lys Leu Leu Tyr Ile Phe Lys Gln Pro Phe Met Arg Pro
340 345 350
Val Gln Thr Thr Gln Glu Glu Asp Gly Cys Ser Cys Arg Phe Pro Glu
355 360 365
Glu Glu Glu Gly Gly Cys Glu Leu Arg Val Lys Phe Ser Arg Ser Ala
370 375 380
Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu
385 390 395 400
Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly
405 410 415
Arg Asp Pro Glu Met Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu
420 425 430
Gly Leu Tyr Asn Glu Leu Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser
435 440 445
Glu Ile Gly Met Lys Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly
450 455 460
Leu Tyr Gln Gly Leu Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu
465 470 475 480
His Met Gln Ala Leu Pro Pro Arg Ala Lys Arg Ser Gly Ser Gly Ala
485 490 495
Thr Asn Phe Ser Leu Leu Lys Gln Ala Gly Asp Val Glu Glu Asn Pro
500 505 510
Gly Pro Met Glu Thr Leu Leu Gly Leu Leu Ile Leu Trp Leu Gln Leu
515 520 525
Gln Trp Val Ser Ser Lys Gln Glu Val Thr Gln Ile Pro Ala Ala Leu
530 535 540
Ser Val Pro Glu Gly Glu Asn Leu Val Leu Asn Cys Ser Phe Thr Asp
545 550 555 560
Ser Ala Ile Tyr Asn Leu Gln Trp Phe Arg Gln Asp Pro Gly Lys Gly
565 570 575
Leu Thr Ser Leu Leu Leu Ile Gln Ser Ser Gln Arg Glu Gln Thr Ser
580 585 590
Gly Arg Leu Asn Ala Ser Leu Asp Lys Ser Ser Gly Arg Ser Thr Leu
595 600 605
Tyr Ile Ala Ala Ser Gln Pro Gly Asp Ser Ala Thr Tyr Leu Cys Ala
610 615 620
Val Arg Pro Leu Tyr Gly Gly Ser Tyr Ile Pro Thr Phe Gly Arg Gly
625 630 635 640
Thr Ser Leu Ile Val His Pro Tyr Ile Gln Asn Pro Asp Pro Ala Val
645 650 655
Tyr Gln Leu Arg Asp Ser Lys Ser Ser Asp Lys Ser Val Cys Leu Phe
660 665 670
Thr Asp Phe Asp Ser Gln Thr Asn Val Ser Gln Ser Lys Asp Ser Asp
675 680 685
Val Tyr Ile Thr Asp Lys Thr Val Leu Asp Met Arg Ser Met Asp Phe
690 695 700
Lys Ser Asn Ser Ala Val Ala Trp Ser Asn Lys Ser Asp Phe Ala Cys
705 710 715 720
Ala Asn Ala Phe Asn Asn Ser Ile Ile Pro Glu Asp Thr Phe Phe Pro
725 730 735
Ser Pro Glu Ser Ser Gly Ser Pro Lys Phe Trp Val Leu Val Val Val
740 745 750
Gly Gly Val Leu Ala Cys Tyr Ser Leu Leu Val Thr Val Ala Phe Ile
755 760 765
Ile Phe Trp Val Arg Ser Lys Arg Ser Arg Leu Leu His Ser Asp Tyr
770 775 780
Met Asn Met Thr Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr Gln
785 790 795 800
Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala Tyr Arg Ser Lys Arg Gly
805 810 815
Arg Lys Lys Leu Leu Tyr Ile Phe Lys Gln Pro Phe Met Arg Pro Val
820 825 830
Gln Thr Thr Gln Glu Glu Asp Gly Cys Ser Cys Arg Phe Pro Glu Glu
835 840 845
Glu Glu Gly Gly Cys Glu Leu Arg Val Lys Phe Ser Arg Ser Ala Asp
850 855 860
Ala Pro Ala Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn
865 870 875 880
Leu Gly Arg Arg Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg
885 890 895
Asp Pro Glu Met Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly
900 905 910
Leu Tyr Asn Glu Leu Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu
915 920 925
Ile Gly Met Lys Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu
930 935 940
Tyr Gln Gly Leu Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His
945 950 955 960
Met Gln Ala Leu Pro Pro Arg
965
<210> 35
<211> 883
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 35
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Gly Ser Pro Lys Phe Trp Val Leu Val Val
260 265 270
Val Gly Gly Val Leu Ala Cys Tyr Ser Leu Leu Val Thr Val Ala Phe
275 280 285
Ile Ile Phe Trp Val Arg Ser Lys Arg Ser Arg Leu Leu His Ser Asp
290 295 300
Tyr Met Asn Met Thr Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr
305 310 315 320
Gln Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala Tyr Arg Ser Arg Val
325 330 335
Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn
340 345 350
Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val
355 360 365
Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met Gly Gly Lys Pro Arg
370 375 380
Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu Leu Gln Lys Asp Lys
385 390 395 400
Met Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys Gly Glu Arg Arg Arg
405 410 415
Gly Lys Gly His Asp Gly Leu Tyr Gln Gly Leu Ser Thr Ala Thr Lys
420 425 430
Asp Thr Tyr Asp Ala Leu His Met Gln Ala Leu Pro Pro Arg Ala Lys
435 440 445
Arg Ser Gly Ser Gly Ala Thr Asn Phe Ser Leu Leu Lys Gln Ala Gly
450 455 460
Asp Val Glu Glu Asn Pro Gly Pro Met Glu Thr Leu Leu Gly Leu Leu
465 470 475 480
Ile Leu Trp Leu Gln Leu Gln Trp Val Ser Ser Lys Gln Glu Val Thr
485 490 495
Gln Ile Pro Ala Ala Leu Ser Val Pro Glu Gly Glu Asn Leu Val Leu
500 505 510
Asn Cys Ser Phe Thr Asp Ser Ala Ile Tyr Asn Leu Gln Trp Phe Arg
515 520 525
Gln Asp Pro Gly Lys Gly Leu Thr Ser Leu Leu Leu Ile Gln Ser Ser
530 535 540
Gln Arg Glu Gln Thr Ser Gly Arg Leu Asn Ala Ser Leu Asp Lys Ser
545 550 555 560
Ser Gly Arg Ser Thr Leu Tyr Ile Ala Ala Ser Gln Pro Gly Asp Ser
565 570 575
Ala Thr Tyr Leu Cys Ala Val Arg Pro Leu Tyr Gly Gly Ser Tyr Ile
580 585 590
Pro Thr Phe Gly Arg Gly Thr Ser Leu Ile Val His Pro Tyr Ile Gln
595 600 605
Asn Pro Asp Pro Ala Val Tyr Gln Leu Arg Asp Ser Lys Ser Ser Asp
610 615 620
Lys Ser Val Cys Leu Phe Thr Asp Phe Asp Ser Gln Thr Asn Val Ser
625 630 635 640
Gln Ser Lys Asp Ser Asp Val Tyr Ile Thr Asp Lys Thr Val Leu Asp
645 650 655
Met Arg Ser Met Asp Phe Lys Ser Asn Ser Ala Val Ala Trp Ser Asn
660 665 670
Lys Ser Asp Phe Ala Cys Ala Asn Ala Phe Asn Asn Ser Ile Ile Pro
675 680 685
Glu Asp Thr Phe Phe Pro Ser Pro Glu Ser Ser Gly Ser Pro Lys Cys
690 695 700
Asp Ile Tyr Ile Trp Ala Pro Leu Ala Gly Thr Cys Gly Val Leu Leu
705 710 715 720
Leu Ser Leu Val Ile Thr Leu Tyr Cys Lys Arg Gly Arg Lys Lys Leu
725 730 735
Leu Tyr Ile Phe Lys Gln Pro Phe Met Arg Pro Val Gln Thr Thr Gln
740 745 750
Glu Glu Asp Gly Cys Ser Cys Arg Phe Pro Glu Glu Glu Glu Gly Gly
755 760 765
Cys Glu Leu Arg Val Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr
770 775 780
Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg
785 790 795 800
Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met
805 810 815
Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu
820 825 830
Leu Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys
835 840 845
Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu Tyr Gln Gly Leu
850 855 860
Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His Met Gln Ala Leu
865 870 875 880
Pro Pro Arg
<210> 36
<211> 771
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 36
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Gly Ser Pro Lys Phe Trp Val Leu Val Val
260 265 270
Val Gly Gly Val Leu Ala Cys Tyr Ser Leu Leu Val Thr Val Ala Phe
275 280 285
Ile Ile Phe Trp Val Arg Ser Lys Arg Ser Arg Leu Leu His Ser Asp
290 295 300
Tyr Met Asn Met Thr Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr
305 310 315 320
Gln Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala Tyr Arg Ser Arg Val
325 330 335
Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn
340 345 350
Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val
355 360 365
Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met Gly Gly Lys Pro Arg
370 375 380
Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu Leu Gln Lys Asp Lys
385 390 395 400
Met Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys Gly Glu Arg Arg Arg
405 410 415
Gly Lys Gly His Asp Gly Leu Tyr Gln Gly Leu Ser Thr Ala Thr Lys
420 425 430
Asp Thr Tyr Asp Ala Leu His Met Gln Ala Leu Pro Pro Arg Ala Lys
435 440 445
Arg Ser Gly Ser Gly Ala Thr Asn Phe Ser Leu Leu Lys Gln Ala Gly
450 455 460
Asp Val Glu Glu Asn Pro Gly Pro Met Glu Thr Leu Leu Gly Leu Leu
465 470 475 480
Ile Leu Trp Leu Gln Leu Gln Trp Val Ser Ser Lys Gln Glu Val Thr
485 490 495
Gln Ile Pro Ala Ala Leu Ser Val Pro Glu Gly Glu Asn Leu Val Leu
500 505 510
Asn Cys Ser Phe Thr Asp Ser Ala Ile Tyr Asn Leu Gln Trp Phe Arg
515 520 525
Gln Asp Pro Gly Lys Gly Leu Thr Ser Leu Leu Leu Ile Gln Ser Ser
530 535 540
Gln Arg Glu Gln Thr Ser Gly Arg Leu Asn Ala Ser Leu Asp Lys Ser
545 550 555 560
Ser Gly Arg Ser Thr Leu Tyr Ile Ala Ala Ser Gln Pro Gly Asp Ser
565 570 575
Ala Thr Tyr Leu Cys Ala Val Arg Pro Leu Tyr Gly Gly Ser Tyr Ile
580 585 590
Pro Thr Phe Gly Arg Gly Thr Ser Leu Ile Val His Pro Tyr Ile Gln
595 600 605
Asn Pro Asp Pro Ala Val Tyr Gln Leu Arg Asp Ser Lys Ser Ser Asp
610 615 620
Lys Ser Val Cys Leu Phe Thr Asp Phe Asp Ser Gln Thr Asn Val Ser
625 630 635 640
Gln Ser Lys Asp Ser Asp Val Tyr Ile Thr Asp Lys Thr Val Leu Asp
645 650 655
Met Arg Ser Met Asp Phe Lys Ser Asn Ser Ala Val Ala Trp Ser Asn
660 665 670
Lys Ser Asp Phe Ala Cys Ala Asn Ala Phe Asn Asn Ser Ile Ile Pro
675 680 685
Glu Asp Thr Phe Phe Pro Ser Pro Glu Ser Ser Gly Ser Pro Lys Cys
690 695 700
Asp Ile Tyr Ile Trp Ala Pro Leu Ala Gly Thr Cys Gly Val Leu Leu
705 710 715 720
Leu Ser Leu Val Ile Thr Leu Tyr Cys Lys Arg Gly Arg Lys Lys Leu
725 730 735
Leu Tyr Ile Phe Lys Gln Pro Phe Met Arg Pro Val Gln Thr Thr Gln
740 745 750
Glu Glu Asp Gly Cys Ser Cys Arg Phe Pro Glu Glu Glu Glu Gly Gly
755 760 765
Cys Glu Leu
770
<210> 37
<211> 884
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 37
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Gly Ser Pro Lys Phe Trp Val Leu Val Val
260 265 270
Val Gly Gly Val Leu Ala Cys Tyr Ser Leu Leu Val Thr Val Ala Phe
275 280 285
Ile Ile Phe Trp Val Arg Ser Lys Arg Ser Arg Leu Leu His Ser Asp
290 295 300
Tyr Met Asn Met Thr Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr
305 310 315 320
Gln Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala Tyr Arg Ser Arg Val
325 330 335
Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn
340 345 350
Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val
355 360 365
Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met Gly Gly Lys Pro Arg
370 375 380
Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu Leu Gln Lys Asp Lys
385 390 395 400
Met Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys Gly Glu Arg Arg Arg
405 410 415
Gly Lys Gly His Asp Gly Leu Tyr Gln Gly Leu Ser Thr Ala Thr Lys
420 425 430
Asp Thr Tyr Asp Ala Leu His Met Gln Ala Leu Pro Pro Arg Ala Lys
435 440 445
Arg Ser Gly Ser Gly Ala Thr Asn Phe Ser Leu Leu Lys Gln Ala Gly
450 455 460
Asp Val Glu Glu Asn Pro Gly Pro Met Glu Thr Leu Leu Gly Leu Leu
465 470 475 480
Ile Leu Trp Leu Gln Leu Gln Trp Val Ser Ser Lys Gln Glu Val Thr
485 490 495
Gln Ile Pro Ala Ala Leu Ser Val Pro Glu Gly Glu Asn Leu Val Leu
500 505 510
Asn Cys Ser Phe Thr Asp Ser Ala Ile Tyr Asn Leu Gln Trp Phe Arg
515 520 525
Gln Asp Pro Gly Lys Gly Leu Thr Ser Leu Leu Leu Ile Gln Ser Ser
530 535 540
Gln Arg Glu Gln Thr Ser Gly Arg Leu Asn Ala Ser Leu Asp Lys Ser
545 550 555 560
Ser Gly Arg Ser Thr Leu Tyr Ile Ala Ala Ser Gln Pro Gly Asp Ser
565 570 575
Ala Thr Tyr Leu Cys Ala Val Arg Pro Leu Tyr Gly Gly Ser Tyr Ile
580 585 590
Pro Thr Phe Gly Arg Gly Thr Ser Leu Ile Val His Pro Tyr Ile Gln
595 600 605
Asn Pro Asp Pro Ala Val Tyr Gln Leu Arg Asp Ser Lys Ser Ser Asp
610 615 620
Lys Ser Val Cys Leu Phe Thr Asp Phe Asp Ser Gln Thr Asn Val Ser
625 630 635 640
Gln Ser Lys Asp Ser Asp Val Tyr Ile Thr Asp Lys Thr Val Leu Asp
645 650 655
Met Arg Ser Met Asp Phe Lys Ser Asn Ser Ala Val Ala Trp Ser Asn
660 665 670
Lys Ser Asp Phe Ala Cys Ala Asn Ala Phe Asn Asn Ser Ile Ile Pro
675 680 685
Glu Asp Thr Phe Phe Pro Ser Pro Glu Ser Ser Gly Ser Pro Lys Phe
690 695 700
Trp Val Leu Val Val Val Gly Gly Val Leu Ala Cys Tyr Ser Leu Leu
705 710 715 720
Val Thr Val Ala Phe Ile Ile Phe Trp Val Lys Arg Gly Arg Lys Lys
725 730 735
Leu Leu Tyr Ile Phe Lys Gln Pro Phe Met Arg Pro Val Gln Thr Thr
740 745 750
Gln Glu Glu Asp Gly Cys Ser Cys Arg Phe Pro Glu Glu Glu Glu Gly
755 760 765
Gly Cys Glu Leu Arg Val Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala
770 775 780
Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg
785 790 795 800
Arg Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu
805 810 815
Met Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn
820 825 830
Glu Leu Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile Gly Met
835 840 845
Lys Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu Tyr Gln Gly
850 855 860
Leu Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His Met Gln Ala
865 870 875 880
Leu Pro Pro Arg
<210> 38
<211> 882
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 38
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Gly Ser Pro Lys Cys Asp Ile Tyr Ile Trp
260 265 270
Ala Pro Leu Ala Gly Thr Cys Gly Val Leu Leu Leu Ser Leu Val Ile
275 280 285
Thr Leu Tyr Cys Arg Ser Lys Arg Ser Arg Leu Leu His Ser Asp Tyr
290 295 300
Met Asn Met Thr Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr Gln
305 310 315 320
Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala Tyr Arg Ser Arg Val Lys
325 330 335
Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn Gln
340 345 350
Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val Leu
355 360 365
Asp Lys Arg Arg Gly Arg Asp Pro Glu Met Gly Gly Lys Pro Arg Arg
370 375 380
Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu Leu Gln Lys Asp Lys Met
385 390 395 400
Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys Gly Glu Arg Arg Arg Gly
405 410 415
Lys Gly His Asp Gly Leu Tyr Gln Gly Leu Ser Thr Ala Thr Lys Asp
420 425 430
Thr Tyr Asp Ala Leu His Met Gln Ala Leu Pro Pro Arg Ala Lys Arg
435 440 445
Ser Gly Ser Gly Ala Thr Asn Phe Ser Leu Leu Lys Gln Ala Gly Asp
450 455 460
Val Glu Glu Asn Pro Gly Pro Met Glu Thr Leu Leu Gly Leu Leu Ile
465 470 475 480
Leu Trp Leu Gln Leu Gln Trp Val Ser Ser Lys Gln Glu Val Thr Gln
485 490 495
Ile Pro Ala Ala Leu Ser Val Pro Glu Gly Glu Asn Leu Val Leu Asn
500 505 510
Cys Ser Phe Thr Asp Ser Ala Ile Tyr Asn Leu Gln Trp Phe Arg Gln
515 520 525
Asp Pro Gly Lys Gly Leu Thr Ser Leu Leu Leu Ile Gln Ser Ser Gln
530 535 540
Arg Glu Gln Thr Ser Gly Arg Leu Asn Ala Ser Leu Asp Lys Ser Ser
545 550 555 560
Gly Arg Ser Thr Leu Tyr Ile Ala Ala Ser Gln Pro Gly Asp Ser Ala
565 570 575
Thr Tyr Leu Cys Ala Val Arg Pro Leu Tyr Gly Gly Ser Tyr Ile Pro
580 585 590
Thr Phe Gly Arg Gly Thr Ser Leu Ile Val His Pro Tyr Ile Gln Asn
595 600 605
Pro Asp Pro Ala Val Tyr Gln Leu Arg Asp Ser Lys Ser Ser Asp Lys
610 615 620
Ser Val Cys Leu Phe Thr Asp Phe Asp Ser Gln Thr Asn Val Ser Gln
625 630 635 640
Ser Lys Asp Ser Asp Val Tyr Ile Thr Asp Lys Thr Val Leu Asp Met
645 650 655
Arg Ser Met Asp Phe Lys Ser Asn Ser Ala Val Ala Trp Ser Asn Lys
660 665 670
Ser Asp Phe Ala Cys Ala Asn Ala Phe Asn Asn Ser Ile Ile Pro Glu
675 680 685
Asp Thr Phe Phe Pro Ser Pro Glu Ser Ser Gly Ser Pro Lys Cys Asp
690 695 700
Ile Tyr Ile Trp Ala Pro Leu Ala Gly Thr Cys Gly Val Leu Leu Leu
705 710 715 720
Ser Leu Val Ile Thr Leu Tyr Cys Lys Arg Gly Arg Lys Lys Leu Leu
725 730 735
Tyr Ile Phe Lys Gln Pro Phe Met Arg Pro Val Gln Thr Thr Gln Glu
740 745 750
Glu Asp Gly Cys Ser Cys Arg Phe Pro Glu Glu Glu Glu Gly Gly Cys
755 760 765
Glu Leu Arg Val Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln
770 775 780
Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu
785 790 795 800
Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met Gly
805 810 815
Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu Leu
820 825 830
Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys Gly
835 840 845
Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu Tyr Gln Gly Leu Ser
850 855 860
Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His Met Gln Ala Leu Pro
865 870 875 880
Pro Arg
<210> 39
<211> 965
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 39
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Gly Ser Pro Lys Cys Asp Ile Tyr Ile Trp
260 265 270
Ala Pro Leu Ala Gly Thr Cys Gly Val Leu Leu Leu Ser Leu Val Ile
275 280 285
Thr Leu Tyr Cys Arg Ser Lys Arg Ser Arg Leu Leu His Ser Asp Tyr
290 295 300
Met Asn Met Thr Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr Gln
305 310 315 320
Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala Tyr Arg Ser Arg Ser Lys
325 330 335
Arg Ser Arg Leu Leu His Ser Asp Tyr Met Asn Met Thr Pro Arg Arg
340 345 350
Pro Gly Pro Thr Arg Lys His Tyr Gln Pro Tyr Ala Pro Pro Arg Asp
355 360 365
Phe Ala Ala Tyr Arg Ser Arg Val Lys Phe Ser Arg Ser Ala Asp Ala
370 375 380
Pro Ala Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu
385 390 395 400
Gly Arg Arg Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg Asp
405 410 415
Pro Glu Met Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu
420 425 430
Tyr Asn Glu Leu Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile
435 440 445
Gly Met Lys Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu Tyr
450 455 460
Gln Gly Leu Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His Met
465 470 475 480
Gln Ala Leu Pro Pro Arg Ala Lys Arg Ser Gly Ser Gly Ala Thr Asn
485 490 495
Phe Ser Leu Leu Lys Gln Ala Gly Asp Val Glu Glu Asn Pro Gly Pro
500 505 510
Met Glu Thr Leu Leu Gly Leu Leu Ile Leu Trp Leu Gln Leu Gln Trp
515 520 525
Val Ser Ser Lys Gln Glu Val Thr Gln Ile Pro Ala Ala Leu Ser Val
530 535 540
Pro Glu Gly Glu Asn Leu Val Leu Asn Cys Ser Phe Thr Asp Ser Ala
545 550 555 560
Ile Tyr Asn Leu Gln Trp Phe Arg Gln Asp Pro Gly Lys Gly Leu Thr
565 570 575
Ser Leu Leu Leu Ile Gln Ser Ser Gln Arg Glu Gln Thr Ser Gly Arg
580 585 590
Leu Asn Ala Ser Leu Asp Lys Ser Ser Gly Arg Ser Thr Leu Tyr Ile
595 600 605
Ala Ala Ser Gln Pro Gly Asp Ser Ala Thr Tyr Leu Cys Ala Val Arg
610 615 620
Pro Leu Tyr Gly Gly Ser Tyr Ile Pro Thr Phe Gly Arg Gly Thr Ser
625 630 635 640
Leu Ile Val His Pro Tyr Ile Gln Asn Pro Asp Pro Ala Val Tyr Gln
645 650 655
Leu Arg Asp Ser Lys Ser Ser Asp Lys Ser Val Cys Leu Phe Thr Asp
660 665 670
Phe Asp Ser Gln Thr Asn Val Ser Gln Ser Lys Asp Ser Asp Val Tyr
675 680 685
Ile Thr Asp Lys Thr Val Leu Asp Met Arg Ser Met Asp Phe Lys Ser
690 695 700
Asn Ser Ala Val Ala Trp Ser Asn Lys Ser Asp Phe Ala Cys Ala Asn
705 710 715 720
Ala Phe Asn Asn Ser Ile Ile Pro Glu Asp Thr Phe Phe Pro Ser Pro
725 730 735
Glu Ser Ser Gly Ser Pro Lys Cys Asp Ile Tyr Ile Trp Ala Pro Leu
740 745 750
Ala Gly Thr Cys Gly Val Leu Leu Leu Ser Leu Val Ile Thr Leu Tyr
755 760 765
Cys Lys Arg Gly Arg Lys Lys Leu Leu Tyr Ile Phe Lys Gln Pro Phe
770 775 780
Met Arg Pro Val Gln Thr Thr Gln Glu Glu Asp Gly Cys Ser Cys Arg
785 790 795 800
Phe Pro Glu Glu Glu Glu Gly Gly Cys Glu Leu Lys Arg Gly Arg Lys
805 810 815
Lys Leu Leu Tyr Ile Phe Lys Gln Pro Phe Met Arg Pro Val Gln Thr
820 825 830
Thr Gln Glu Glu Asp Gly Cys Ser Cys Arg Phe Pro Glu Glu Glu Glu
835 840 845
Gly Gly Cys Glu Leu Arg Val Lys Phe Ser Arg Ser Ala Asp Ala Pro
850 855 860
Ala Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu Gly
865 870 875 880
Arg Arg Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg Asp Pro
885 890 895
Glu Met Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu Tyr
900 905 910
Asn Glu Leu Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile Gly
915 920 925
Met Lys Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu Tyr Gln
930 935 940
Gly Leu Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His Met Gln
945 950 955 960
Ala Leu Pro Pro Arg
965
<210> 40
<211> 1059
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 40
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Gly Ser Pro Lys Ala Lys Pro Thr Thr Thr
260 265 270
Pro Ala Pro Arg Pro Pro Thr Pro Ala Pro Thr Ile Ala Ser Gln Pro
275 280 285
Leu Ser Leu Arg Pro Glu Ala Cys Arg Pro Ala Ala Gly Gly Ala Val
290 295 300
His Thr Arg Gly Leu Asp Phe Ala Pro Arg Lys Ile Glu Val Met Tyr
305 310 315 320
Pro Pro Pro Tyr Leu Asp Asn Glu Lys Ser Asn Gly Thr Ile Ile His
325 330 335
Val Lys Gly Lys His Leu Cys Pro Ser Pro Leu Phe Pro Gly Pro Ser
340 345 350
Lys Pro Phe Trp Val Leu Val Val Val Gly Gly Val Leu Ala Cys Tyr
355 360 365
Ser Leu Leu Val Thr Val Ala Phe Ile Ile Phe Trp Val Arg Ser Lys
370 375 380
Arg Ser Arg Leu Leu His Ser Asp Tyr Met Asn Met Thr Pro Arg Arg
385 390 395 400
Pro Gly Pro Thr Arg Lys His Tyr Gln Pro Tyr Ala Pro Pro Arg Asp
405 410 415
Phe Ala Ala Tyr Arg Ser Arg Val Lys Phe Ser Arg Ser Ala Asp Ala
420 425 430
Pro Ala Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu
435 440 445
Gly Arg Arg Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg Asp
450 455 460
Pro Glu Met Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu
465 470 475 480
Tyr Asn Glu Leu Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile
485 490 495
Gly Met Lys Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu Tyr
500 505 510
Gln Gly Leu Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His Met
515 520 525
Gln Ala Leu Pro Pro Arg Ala Lys Arg Ser Gly Ser Gly Ala Thr Asn
530 535 540
Phe Ser Leu Leu Lys Gln Ala Gly Asp Val Glu Glu Asn Pro Gly Pro
545 550 555 560
Met Glu Thr Leu Leu Gly Leu Leu Ile Leu Trp Leu Gln Leu Gln Trp
565 570 575
Val Ser Ser Lys Gln Glu Val Thr Gln Ile Pro Ala Ala Leu Ser Val
580 585 590
Pro Glu Gly Glu Asn Leu Val Leu Asn Cys Ser Phe Thr Asp Ser Ala
595 600 605
Ile Tyr Asn Leu Gln Trp Phe Arg Gln Asp Pro Gly Lys Gly Leu Thr
610 615 620
Ser Leu Leu Leu Ile Gln Ser Ser Gln Arg Glu Gln Thr Ser Gly Arg
625 630 635 640
Leu Asn Ala Ser Leu Asp Lys Ser Ser Gly Arg Ser Thr Leu Tyr Ile
645 650 655
Ala Ala Ser Gln Pro Gly Asp Ser Ala Thr Tyr Leu Cys Ala Val Arg
660 665 670
Pro Leu Tyr Gly Gly Ser Tyr Ile Pro Thr Phe Gly Arg Gly Thr Ser
675 680 685
Leu Ile Val His Pro Tyr Ile Gln Asn Pro Asp Pro Ala Val Tyr Gln
690 695 700
Leu Arg Asp Ser Lys Ser Ser Asp Lys Ser Val Cys Leu Phe Thr Asp
705 710 715 720
Phe Asp Ser Gln Thr Asn Val Ser Gln Ser Lys Asp Ser Asp Val Tyr
725 730 735
Ile Thr Asp Lys Thr Val Leu Asp Met Arg Ser Met Asp Phe Lys Ser
740 745 750
Asn Ser Ala Val Ala Trp Ser Asn Lys Ser Asp Phe Ala Cys Ala Asn
755 760 765
Ala Phe Asn Asn Ser Ile Ile Pro Glu Asp Thr Phe Phe Pro Ser Pro
770 775 780
Glu Ser Ser Gly Ser Pro Lys Ala Lys Pro Thr Thr Thr Pro Ala Pro
785 790 795 800
Arg Pro Pro Thr Pro Ala Pro Thr Ile Ala Ser Gln Pro Leu Ser Leu
805 810 815
Arg Pro Glu Ala Cys Arg Pro Ala Ala Gly Gly Ala Val His Thr Arg
820 825 830
Gly Leu Asp Phe Ala Pro Arg Lys Ile Glu Val Met Tyr Pro Pro Pro
835 840 845
Tyr Leu Asp Asn Glu Lys Ser Asn Gly Thr Ile Ile His Val Lys Gly
850 855 860
Lys His Leu Cys Pro Ser Pro Leu Phe Pro Gly Pro Ser Lys Pro Phe
865 870 875 880
Trp Val Leu Val Val Val Gly Gly Val Leu Ala Cys Tyr Ser Leu Leu
885 890 895
Val Thr Val Ala Phe Ile Ile Phe Trp Val Arg Ser Lys Arg Ser Arg
900 905 910
Leu Leu His Ser Asp Tyr Met Asn Met Thr Pro Arg Arg Pro Gly Pro
915 920 925
Thr Arg Lys His Tyr Gln Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala
930 935 940
Tyr Arg Ser Arg Val Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr
945 950 955 960
Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg
965 970 975
Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met
980 985 990
Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu
995 1000 1005
Leu Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile Gly Met
1010 1015 1020
Lys Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu Tyr Gln
1025 1030 1035
Gly Leu Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His Met
1040 1045 1050
Gln Ala Leu Pro Pro Arg
1055
<210> 41
<211> 859
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 41
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Gly Ser Pro Lys Ala Lys Pro Thr Thr Thr
260 265 270
Pro Ala Pro Arg Pro Pro Thr Pro Ala Pro Thr Ile Ala Ser Gln Pro
275 280 285
Leu Ser Leu Arg Pro Glu Ala Cys Arg Pro Ala Ala Gly Gly Ala Val
290 295 300
His Thr Arg Gly Leu Asp Phe Ala Pro Arg Lys Ile Glu Val Met Tyr
305 310 315 320
Pro Pro Pro Tyr Leu Asp Asn Glu Lys Ser Asn Gly Thr Ile Ile His
325 330 335
Val Lys Gly Lys His Leu Cys Pro Ser Pro Leu Phe Pro Gly Pro Ser
340 345 350
Lys Pro Phe Trp Val Leu Val Val Val Gly Gly Val Leu Ala Cys Tyr
355 360 365
Ser Leu Leu Val Thr Val Ala Phe Ile Ile Phe Trp Val Arg Ser Lys
370 375 380
Arg Ser Arg Leu Leu His Ser Asp Tyr Met Asn Met Thr Pro Arg Arg
385 390 395 400
Pro Gly Pro Thr Arg Lys His Tyr Gln Pro Tyr Ala Pro Pro Arg Asp
405 410 415
Phe Ala Ala Tyr Arg Ser Arg Val Lys Phe Ser Arg Ser Ala Asp Ala
420 425 430
Pro Ala Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu
435 440 445
Gly Arg Arg Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg Asp
450 455 460
Pro Glu Met Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu
465 470 475 480
Tyr Asn Glu Leu Gln Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile
485 490 495
Gly Met Lys Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu Tyr
500 505 510
Gln Gly Leu Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His Met
515 520 525
Gln Ala Leu Pro Pro Arg Ala Lys Arg Ser Gly Ser Gly Ala Thr Asn
530 535 540
Phe Ser Leu Leu Lys Gln Ala Gly Asp Val Glu Glu Asn Pro Gly Pro
545 550 555 560
Met Glu Thr Leu Leu Gly Leu Leu Ile Leu Trp Leu Gln Leu Gln Trp
565 570 575
Val Ser Ser Lys Gln Glu Val Thr Gln Ile Pro Ala Ala Leu Ser Val
580 585 590
Pro Glu Gly Glu Asn Leu Val Leu Asn Cys Ser Phe Thr Asp Ser Ala
595 600 605
Ile Tyr Asn Leu Gln Trp Phe Arg Gln Asp Pro Gly Lys Gly Leu Thr
610 615 620
Ser Leu Leu Leu Ile Gln Ser Ser Gln Arg Glu Gln Thr Ser Gly Arg
625 630 635 640
Leu Asn Ala Ser Leu Asp Lys Ser Ser Gly Arg Ser Thr Leu Tyr Ile
645 650 655
Ala Ala Ser Gln Pro Gly Asp Ser Ala Thr Tyr Leu Cys Ala Val Arg
660 665 670
Pro Leu Tyr Gly Gly Ser Tyr Ile Pro Thr Phe Gly Arg Gly Thr Ser
675 680 685
Leu Ile Val His Pro Tyr Ile Gln Asn Pro Asp Pro Ala Val Tyr Gln
690 695 700
Leu Arg Asp Ser Lys Ser Ser Asp Lys Ser Val Cys Leu Phe Thr Asp
705 710 715 720
Phe Asp Ser Gln Thr Asn Val Ser Gln Ser Lys Asp Ser Asp Val Tyr
725 730 735
Ile Thr Asp Lys Thr Val Leu Asp Met Arg Ser Met Asp Phe Lys Ser
740 745 750
Asn Ser Ala Val Ala Trp Ser Asn Lys Ser Asp Phe Ala Cys Ala Asn
755 760 765
Ala Phe Asn Asn Ser Ile Ile Pro Glu Asp Thr Phe Phe Pro Ser Pro
770 775 780
Glu Ser Ser Gly Ser Pro Lys Cys Asp Ile Tyr Ile Trp Ala Pro Leu
785 790 795 800
Ala Gly Thr Cys Gly Val Leu Leu Leu Ser Leu Val Ile Thr Leu Tyr
805 810 815
Cys Lys Arg Gly Arg Lys Lys Leu Leu Tyr Ile Phe Lys Gln Pro Phe
820 825 830
Met Arg Pro Val Gln Thr Thr Gln Glu Glu Asp Gly Cys Ser Cys Arg
835 840 845
Phe Pro Glu Glu Glu Glu Gly Gly Cys Glu Leu
850 855
<210> 42
<211> 120
<212> DNA
<213> Intelligent people
<400> 42
aaaattgaag ttatgtatcc tcctccttac ctagacaatg agaagagcaa tggaaccatt 60
atccatgtga aagggaaaca cctttgtcca agtcccctat ttcccggacc ttctaagccc 120
<210> 43
<211> 120
<212> DNA
<213> Intelligent people
<400> 43
aagatcgagg taatgtaccc accgccctat cttgataacg aaaaatctaa cggtacaata 60
attcacgtca agggcaagca tttgtgccct tccccgttgt tcccgggccc aagcaaaccg 120
<210> 44
<211> 81
<212> DNA
<213> Intelligent people
<400> 44
ttttgggtgc tggtggtggt tggtggagtc ctggcttgct atagcttgct agtaacagtg 60
gcctttatta ttttctgggt g 81
<210> 45
<211> 81
<212> DNA
<213> Intelligent people
<400> 45
ttctgggttc tcgtcgtcgt gggaggtgtg ttagcatgtt actctctctt ggttactgtc 60
gctttcataa tcttttgggt c 81
<210> 46
<211> 123
<212> DNA
<213> Intelligent people
<400> 46
aggagtaaga ggagcaggct cctgcacagt gactacatga acatgactcc ccgccgcccc 60
gggcccaccc gcaagcatta ccagccctat gccccaccac gcgacttcgc agcctatcgc 120
tcc 123
<210> 47
<211> 123
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 47
cgctcaaaac gctctcgctt gttacattcc gattatatga atatgacacc taggagacct 60
ggcccgacta ggaaacacta tcaaccttac gcacctccca gagattttgc tgcttacagg 120
agt 123
<210> 48
<211> 126
<212> DNA
<213> Intelligent people
<400> 48
aaacggggca gaaagaaact cctgtatata ttcaaacaac catttatgag accagtacaa 60
actactcaag aggaagatgg ctgtagctgc cgatttccag aagaagaaga aggaggatgt 120
gaactg 126
<210> 49
<211> 126
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 49
aagagagggc gtaaaaagct gctctacatc tttaagcagc ctttcatgcg tcctgttcag 60
acaacacagg aagaggacgg atgctcttgc aggttccctg aggaggagga gggtggttgc 120
gagctc 126
<210> 50
<211> 138
<212> DNA
<213> Intelligent people
<400> 50
gctaagccca ccacgacgcc agcgccgcga ccaccaacac cggcgcccac catcgcgtcg 60
cagcccctgt ccctgcgccc agaggcgtgc cggccagcgg cggggggcgc agtgcacacg 120
agggggctgg acttcgcc 138
<210> 51
<211> 138
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 51
gctaagccca ctactacccc agctcccagg cctcccacac ctgccccaac aatcgccagc 60
cagccactgt cccttaggcc cgaggcctgt aggcccgccg ccggaggagc cgtgcacacc 120
cgcggactgg attttgct 138
<210> 52
<211> 78
<212> DNA
<213> Intelligent people
<400> 52
tgtgatatct acatctgggc gcccttggcc gggacttgtg gggtccttct cctgtcactg 60
gttatcaccc tttactgc 78
<210> 53
<211> 78
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 53
tgcgacattt atatttgggc ccctctcgct ggcacatgcg gcgtgttgtt gctcagcctc 60
gtgattacac tttattgt 78
<210> 54
<211> 2652
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 54
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagacggct ctcctaagtg cgacatttat atttgggccc ctctcgctgg cacatgcggc 840
gtgttgttgc tcagcctcgt gattacactt tattgtaaga gagggcgtaa aaagctgctc 900
tacatcttta agcagccttt catgcgtcct gttcagacaa cacaggaaga ggacggatgc 960
tcttgcaggt tccctgagga ggaggagggt ggttgcgagc tccgggtcaa attttcacgc 1020
tccgctgatg ctcctgccta tcaacaaggg caaaatcaat tgtacaatga attgaacttg 1080
ggtagaaggg aagaatatga cgtgctcgat aaacggaggg ggagagatcc agaaatgggc 1140
ggtaaaccac ggcgcaaaaa tccacaagag ggattgtata acgagctcca aaaggacaaa 1200
atggcagaag cttattcaga aataggaatg aagggggaaa ggagacgagg taaaggtcat 1260
gacggattgt atcaaggatt gtcaaccgct actaaagata catatgatgc tttgcatatg 1320
caagctttgc ctcccagagc caagcggtct gggtctgggg ccaccaactt cagcctgctg 1380
aagcaggccg gcgacgtgga ggagaacccc ggccccatgg agaccctctt gggcctgctt 1440
atcctttggc tgcagctgca atgggtgagc agcaaacagg aggtgacgca gattcctgca 1500
gctctgagtg tcccagaagg agaaaacttg gttctcaact gcagtttcac tgatagcgct 1560
atttacaacc tccagtggtt taggcaggac cctgggaaag gtctcacatc tctgttgctt 1620
attcagtcaa gtcagagaga gcaaacaagt ggaagactta atgcctcgct ggataaatca 1680
tcaggacgta gtactttata cattgcagct tctcagcctg gtgactcagc cacctacctc 1740
tgtgctgtga ggcccctgta cggaggaagc tacataccta catttggaag aggaaccagc 1800
cttattgttc atccgtatat ccagaaccct gaccctgccg tgtaccagct gagagactct 1860
aaatccagtg acaagtctgt ctgcctattc accgattttg attctcaaac aaatgtgtca 1920
caaagtaagg attctgatgt gtatatcaca gacaaaactg tgctagacat gaggtctatg 1980
gacttcaaga gcaacagtgc tgtggcctgg agcaacaaat ctgactttgc atgtgcaaac 2040
gccttcaaca acagcattat tccagaagac accttcttcc ccagcccaga aagttccggc 2100
tccccaaaat gtgatatcta catctgggcg cccttggccg ggacttgtgg ggtccttctc 2160
ctgtcactgg ttatcaccct ttactgcaaa cggggcagaa agaaactcct gtatatattc 2220
aaacaaccat ttatgagacc agtacaaact actcaagagg aagatggctg tagctgccga 2280
tttccagaag aagaagaagg aggatgtgaa ctgagagtga agttcagcag gagcgcagac 2340
gcccccgcgt accagcaggg ccagaaccag ctctataacg agctcaatct aggacgaaga 2400
gaggagtacg atgttttgga caagagacgt ggccgggacc ctgagatggg gggaaagccg 2460
agaaggaaga accctcagga aggcctgtac aatgaactgc agaaagataa gatggcggag 2520
gcctacagtg agattgggat gaaaggcgag cgccggaggg gcaaggggca cgatggcctt 2580
taccagggtc tcagtacagc caccaaggac acctacgacg cccttcacat gcaggccctg 2640
ccccctcgct aa 2652
<210> 55
<211> 2658
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 55
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagacggct ctcctaagtt ctgggttctc gtcgtcgtgg gaggtgtgtt agcatgttac 840
tctctcttgg ttactgtcgc tttcataatc ttttgggtca agagagggcg taaaaagctg 900
ctctacatct ttaagcagcc tttcatgcgt cctgttcaga caacacagga agaggacgga 960
tgctcttgca ggttccctga ggaggaggag ggtggttgcg agctccgggt caaattttca 1020
cgctccgctg atgctcctgc ctatcaacaa gggcaaaatc aattgtacaa tgaattgaac 1080
ttgggtagaa gggaagaata tgacgtgctc gataaacgga gggggagaga tccagaaatg 1140
ggcggtaaac cacggcgcaa aaatccacaa gagggattgt ataacgagct ccaaaaggac 1200
aaaatggcag aagcttattc agaaatagga atgaaggggg aaaggagacg aggtaaaggt 1260
catgacggat tgtatcaagg attgtcaacc gctactaaag atacatatga tgctttgcat 1320
atgcaagctt tgcctcccag agccaagcgg tctgggtctg gggccaccaa cttcagcctg 1380
ctgaagcagg ccggcgacgt ggaggagaac cccggcccca tggagaccct cttgggcctg 1440
cttatccttt ggctgcagct gcaatgggtg agcagcaaac aggaggtgac gcagattcct 1500
gcagctctga gtgtcccaga aggagaaaac ttggttctca actgcagttt cactgatagc 1560
gctatttaca acctccagtg gtttaggcag gaccctggga aaggtctcac atctctgttg 1620
cttattcagt caagtcagag agagcaaaca agtggaagac ttaatgcctc gctggataaa 1680
tcatcaggac gtagtacttt atacattgca gcttctcagc ctggtgactc agccacctac 1740
ctctgtgctg tgaggcccct gtacggagga agctacatac ctacatttgg aagaggaacc 1800
agccttattg ttcatccgta tatccagaac cctgaccctg ccgtgtacca gctgagagac 1860
tctaaatcca gtgacaagtc tgtctgccta ttcaccgatt ttgattctca aacaaatgtg 1920
tcacaaagta aggattctga tgtgtatatc acagacaaaa ctgtgctaga catgaggtct 1980
atggacttca agagcaacag tgctgtggcc tggagcaaca aatctgactt tgcatgtgca 2040
aacgccttca acaacagcat tattccagaa gacaccttct tccccagccc agaaagttcc 2100
ggctccccaa aattttgggt gctggtggtg gttggtggag tcctggcttg ctatagcttg 2160
ctagtaacag tggcctttat tattttctgg gtgaaacggg gcagaaagaa actcctgtat 2220
atattcaaac aaccatttat gagaccagta caaactactc aagaggaaga tggctgtagc 2280
tgccgatttc cagaagaaga agaaggagga tgtgaactga gagtgaagtt cagcaggagc 2340
gcagacgccc ccgcgtacca gcagggccag aaccagctct ataacgagct caatctagga 2400
cgaagagagg agtacgatgt tttggacaag agacgtggcc gggaccctga gatgggggga 2460
aagccgagaa ggaagaaccc tcaggaaggc ctgtacaatg aactgcagaa agataagatg 2520
gcggaggcct acagtgagat tgggatgaaa ggcgagcgcc ggaggggcaa ggggcacgat 2580
ggcctttacc agggtctcag tacagccacc aaggacacct acgacgccct tcacatgcag 2640
gccctgcccc ctcgctaa 2658
<210> 56
<211> 2904
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 56
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagacggct ctcctaagtt ctgggttctc gtcgtcgtgg gaggtgtgtt agcatgttac 840
tctctcttgg ttactgtcgc tttcataatc ttttgggtcc gctcaaaacg ctctcgcttg 900
ttacattccg attatatgaa tatgacacct aggagacctg gcccgactag gaaacactat 960
caaccttacg cacctcccag agattttgct gcttacagga gtaagagagg gcgtaaaaag 1020
ctgctctaca tctttaagca gcctttcatg cgtcctgttc agacaacaca ggaagaggac 1080
ggatgctctt gcaggttccc tgaggaggag gagggtggtt gcgagctccg ggtcaaattt 1140
tcacgctccg ctgatgctcc tgcctatcaa caagggcaaa atcaattgta caatgaattg 1200
aacttgggta gaagggaaga atatgacgtg ctcgataaac ggagggggag agatccagaa 1260
atgggcggta aaccacggcg caaaaatcca caagagggat tgtataacga gctccaaaag 1320
gacaaaatgg cagaagctta ttcagaaata ggaatgaagg gggaaaggag acgaggtaaa 1380
ggtcatgacg gattgtatca aggattgtca accgctacta aagatacata tgatgctttg 1440
catatgcaag ctttgcctcc cagagccaag cggtctgggt ctggggccac caacttcagc 1500
ctgctgaagc aggccggcga cgtggaggag aaccccggcc ccatggagac cctcttgggc 1560
ctgcttatcc tttggctgca gctgcaatgg gtgagcagca aacaggaggt gacgcagatt 1620
cctgcagctc tgagtgtccc agaaggagaa aacttggttc tcaactgcag tttcactgat 1680
agcgctattt acaacctcca gtggtttagg caggaccctg ggaaaggtct cacatctctg 1740
ttgcttattc agtcaagtca gagagagcaa acaagtggaa gacttaatgc ctcgctggat 1800
aaatcatcag gacgtagtac tttatacatt gcagcttctc agcctggtga ctcagccacc 1860
tacctctgtg ctgtgaggcc cctgtacgga ggaagctaca tacctacatt tggaagagga 1920
accagcctta ttgttcatcc gtatatccag aaccctgacc ctgccgtgta ccagctgaga 1980
gactctaaat ccagtgacaa gtctgtctgc ctattcaccg attttgattc tcaaacaaat 2040
gtgtcacaaa gtaaggattc tgatgtgtat atcacagaca aaactgtgct agacatgagg 2100
tctatggact tcaagagcaa cagtgctgtg gcctggagca acaaatctga ctttgcatgt 2160
gcaaacgcct tcaacaacag cattattcca gaagacacct tcttccccag cccagaaagt 2220
tccggctccc caaaattttg ggtgctggtg gtggttggtg gagtcctggc ttgctatagc 2280
ttgctagtaa cagtggcctt tattattttc tgggtgagga gtaagaggag caggctcctg 2340
cacagtgact acatgaacat gactccccgc cgccccgggc ccacccgcaa gcattaccag 2400
ccctatgccc caccacgcga cttcgcagcc tatcgctcca aacggggcag aaagaaactc 2460
ctgtatatat tcaaacaacc atttatgaga ccagtacaaa ctactcaaga ggaagatggc 2520
tgtagctgcc gatttccaga agaagaagaa ggaggatgtg aactgagagt gaagttcagc 2580
aggagcgcag acgcccccgc gtaccagcag ggccagaacc agctctataa cgagctcaat 2640
ctaggacgaa gagaggagta cgatgttttg gacaagagac gtggccggga ccctgagatg 2700
gggggaaagc cgagaaggaa gaaccctcag gaaggcctgt acaatgaact gcagaaagat 2760
aagatggcgg aggcctacag tgagattggg atgaaaggcg agcgccggag gggcaagggg 2820
cacgatggcc tttaccaggg tctcagtaca gccaccaagg acacctacga cgcccttcac 2880
atgcaggccc tgccccctcg ctaa 2904
<210> 57
<211> 2652
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 57
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagacggct ctcctaagtt ctgggttctc gtcgtcgtgg gaggtgtgtt agcatgttac 840
tctctcttgg ttactgtcgc tttcataatc ttttgggtcc gctcaaaacg ctctcgcttg 900
ttacattccg attatatgaa tatgacacct aggagacctg gcccgactag gaaacactat 960
caaccttacg cacctcccag agattttgct gcttacagga gtcgggtcaa attttcacgc 1020
tccgctgatg ctcctgccta tcaacaaggg caaaatcaat tgtacaatga attgaacttg 1080
ggtagaaggg aagaatatga cgtgctcgat aaacggaggg ggagagatcc agaaatgggc 1140
ggtaaaccac ggcgcaaaaa tccacaagag ggattgtata acgagctcca aaaggacaaa 1200
atggcagaag cttattcaga aataggaatg aagggggaaa ggagacgagg taaaggtcat 1260
gacggattgt atcaaggatt gtcaaccgct actaaagata catatgatgc tttgcatatg 1320
caagctttgc ctcccagagc caagcggtct gggtctgggg ccaccaactt cagcctgctg 1380
aagcaggccg gcgacgtgga ggagaacccc ggccccatgg agaccctctt gggcctgctt 1440
atcctttggc tgcagctgca atgggtgagc agcaaacagg aggtgacgca gattcctgca 1500
gctctgagtg tcccagaagg agaaaacttg gttctcaact gcagtttcac tgatagcgct 1560
atttacaacc tccagtggtt taggcaggac cctgggaaag gtctcacatc tctgttgctt 1620
attcagtcaa gtcagagaga gcaaacaagt ggaagactta atgcctcgct ggataaatca 1680
tcaggacgta gtactttata cattgcagct tctcagcctg gtgactcagc cacctacctc 1740
tgtgctgtga ggcccctgta cggaggaagc tacataccta catttggaag aggaaccagc 1800
cttattgttc atccgtatat ccagaaccct gaccctgccg tgtaccagct gagagactct 1860
aaatccagtg acaagtctgt ctgcctattc accgattttg attctcaaac aaatgtgtca 1920
caaagtaagg attctgatgt gtatatcaca gacaaaactg tgctagacat gaggtctatg 1980
gacttcaaga gcaacagtgc tgtggcctgg agcaacaaat ctgactttgc atgtgcaaac 2040
gccttcaaca acagcattat tccagaagac accttcttcc ccagcccaga aagttccggc 2100
tccccaaaat gtgatatcta catctgggcg cccttggccg ggacttgtgg ggtccttctc 2160
ctgtcactgg ttatcaccct ttactgcaaa cggggcagaa agaaactcct gtatatattc 2220
aaacaaccat ttatgagacc agtacaaact actcaagagg aagatggctg tagctgccga 2280
tttccagaag aagaagaagg aggatgtgaa ctgagagtga agttcagcag gagcgcagac 2340
gcccccgcgt accagcaggg ccagaaccag ctctataacg agctcaatct aggacgaaga 2400
gaggagtacg atgttttgga caagagacgt ggccgggacc ctgagatggg gggaaagccg 2460
agaaggaaga accctcagga aggcctgtac aatgaactgc agaaagataa gatggcggag 2520
gcctacagtg agattgggat gaaaggcgag cgccggaggg gcaaggggca cgatggcctt 2580
taccagggtc tcagtacagc caccaaggac acctacgacg cccttcacat gcaggccctg 2640
ccccctcgct aa 2652
<210> 58
<211> 2316
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 58
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagacggct ctcctaagtt ctgggttctc gtcgtcgtgg gaggtgtgtt agcatgttac 840
tctctcttgg ttactgtcgc tttcataatc ttttgggtcc gctcaaaacg ctctcgcttg 900
ttacattccg attatatgaa tatgacacct aggagacctg gcccgactag gaaacactat 960
caaccttacg cacctcccag agattttgct gcttacagga gtcgggtcaa attttcacgc 1020
tccgctgatg ctcctgccta tcaacaaggg caaaatcaat tgtacaatga attgaacttg 1080
ggtagaaggg aagaatatga cgtgctcgat aaacggaggg ggagagatcc agaaatgggc 1140
ggtaaaccac ggcgcaaaaa tccacaagag ggattgtata acgagctcca aaaggacaaa 1200
atggcagaag cttattcaga aataggaatg aagggggaaa ggagacgagg taaaggtcat 1260
gacggattgt atcaaggatt gtcaaccgct actaaagata catatgatgc tttgcatatg 1320
caagctttgc ctcccagagc caagcggtct gggtctgggg ccaccaactt cagcctgctg 1380
aagcaggccg gcgacgtgga ggagaacccc ggccccatgg agaccctctt gggcctgctt 1440
atcctttggc tgcagctgca atgggtgagc agcaaacagg aggtgacgca gattcctgca 1500
gctctgagtg tcccagaagg agaaaacttg gttctcaact gcagtttcac tgatagcgct 1560
atttacaacc tccagtggtt taggcaggac cctgggaaag gtctcacatc tctgttgctt 1620
attcagtcaa gtcagagaga gcaaacaagt ggaagactta atgcctcgct ggataaatca 1680
tcaggacgta gtactttata cattgcagct tctcagcctg gtgactcagc cacctacctc 1740
tgtgctgtga ggcccctgta cggaggaagc tacataccta catttggaag aggaaccagc 1800
cttattgttc atccgtatat ccagaaccct gaccctgccg tgtaccagct gagagactct 1860
aaatccagtg acaagtctgt ctgcctattc accgattttg attctcaaac aaatgtgtca 1920
caaagtaagg attctgatgt gtatatcaca gacaaaactg tgctagacat gaggtctatg 1980
gacttcaaga gcaacagtgc tgtggcctgg agcaacaaat ctgactttgc atgtgcaaac 2040
gccttcaaca acagcattat tccagaagac accttcttcc ccagcccaga aagttccggc 2100
tccccaaaat gtgatatcta catctgggcg cccttggccg ggacttgtgg ggtccttctc 2160
ctgtcactgg ttatcaccct ttactgcaaa cggggcagaa agaaactcct gtatatattc 2220
aaacaaccat ttatgagacc agtacaaact actcaagagg aagatggctg tagctgccga 2280
tttccagaag aagaagaagg aggatgtgaa ctgtaa 2316
<210> 59
<211> 2655
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 59
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagacggct ctcctaagtt ctgggttctc gtcgtcgtgg gaggtgtgtt agcatgttac 840
tctctcttgg ttactgtcgc tttcataatc ttttgggtcc gctcaaaacg ctctcgcttg 900
ttacattccg attatatgaa tatgacacct aggagacctg gcccgactag gaaacactat 960
caaccttacg cacctcccag agattttgct gcttacagga gtcgggtcaa attttcacgc 1020
tccgctgatg ctcctgccta tcaacaaggg caaaatcaat tgtacaatga attgaacttg 1080
ggtagaaggg aagaatatga cgtgctcgat aaacggaggg ggagagatcc agaaatgggc 1140
ggtaaaccac ggcgcaaaaa tccacaagag ggattgtata acgagctcca aaaggacaaa 1200
atggcagaag cttattcaga aataggaatg aagggggaaa ggagacgagg taaaggtcat 1260
gacggattgt atcaaggatt gtcaaccgct actaaagata catatgatgc tttgcatatg 1320
caagctttgc ctcccagagc caagcggtct gggtctgggg ccaccaactt cagcctgctg 1380
aagcaggccg gcgacgtgga ggagaacccc ggccccatgg agaccctctt gggcctgctt 1440
atcctttggc tgcagctgca atgggtgagc agcaaacagg aggtgacgca gattcctgca 1500
gctctgagtg tcccagaagg agaaaacttg gttctcaact gcagtttcac tgatagcgct 1560
atttacaacc tccagtggtt taggcaggac cctgggaaag gtctcacatc tctgttgctt 1620
attcagtcaa gtcagagaga gcaaacaagt ggaagactta atgcctcgct ggataaatca 1680
tcaggacgta gtactttata cattgcagct tctcagcctg gtgactcagc cacctacctc 1740
tgtgctgtga ggcccctgta cggaggaagc tacataccta catttggaag aggaaccagc 1800
cttattgttc atccgtatat ccagaaccct gaccctgccg tgtaccagct gagagactct 1860
aaatccagtg acaagtctgt ctgcctattc accgattttg attctcaaac aaatgtgtca 1920
caaagtaagg attctgatgt gtatatcaca gacaaaactg tgctagacat gaggtctatg 1980
gacttcaaga gcaacagtgc tgtggcctgg agcaacaaat ctgactttgc atgtgcaaac 2040
gccttcaaca acagcattat tccagaagac accttcttcc ccagcccaga aagttccggc 2100
tccccaaaat tttgggtgct ggtggtggtt ggtggagtcc tggcttgcta tagcttgcta 2160
gtaacagtgg cctttattat tttctgggtg aaacggggca gaaagaaact cctgtatata 2220
ttcaaacaac catttatgag accagtacaa actactcaag aggaagatgg ctgtagctgc 2280
cgatttccag aagaagaaga aggaggatgt gaactgagag tgaagttcag caggagcgca 2340
gacgcccccg cgtaccagca gggccagaac cagctctata acgagctcaa tctaggacga 2400
agagaggagt acgatgtttt ggacaagaga cgtggccggg accctgagat ggggggaaag 2460
ccgagaagga agaaccctca ggaaggcctg tacaatgaac tgcagaaaga taagatggcg 2520
gaggcctaca gtgagattgg gatgaaaggc gagcgccgga ggggcaaggg gcacgatggc 2580
ctttaccagg gtctcagtac agccaccaag gacacctacg acgcccttca catgcaggcc 2640
ctgccccctc gctaa 2655
<210> 60
<211> 2649
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 60
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagacggct ctcctaagtg cgacatttat atttgggccc ctctcgctgg cacatgcggc 840
gtgttgttgc tcagcctcgt gattacactt tattgtcgct caaaacgctc tcgcttgtta 900
cattccgatt atatgaatat gacacctagg agacctggcc cgactaggaa acactatcaa 960
ccttacgcac ctcccagaga ttttgctgct tacaggagtc gggtcaaatt ttcacgctcc 1020
gctgatgctc ctgcctatca acaagggcaa aatcaattgt acaatgaatt gaacttgggt 1080
agaagggaag aatatgacgt gctcgataaa cggaggggga gagatccaga aatgggcggt 1140
aaaccacggc gcaaaaatcc acaagaggga ttgtataacg agctccaaaa ggacaaaatg 1200
gcagaagctt attcagaaat aggaatgaag ggggaaagga gacgaggtaa aggtcatgac 1260
ggattgtatc aaggattgtc aaccgctact aaagatacat atgatgcttt gcatatgcaa 1320
gctttgcctc ccagagccaa gcggtctggg tctggggcca ccaacttcag cctgctgaag 1380
caggccggcg acgtggagga gaaccccggc cccatggaga ccctcttggg cctgcttatc 1440
ctttggctgc agctgcaatg ggtgagcagc aaacaggagg tgacgcagat tcctgcagct 1500
ctgagtgtcc cagaaggaga aaacttggtt ctcaactgca gtttcactga tagcgctatt 1560
tacaacctcc agtggtttag gcaggaccct gggaaaggtc tcacatctct gttgcttatt 1620
cagtcaagtc agagagagca aacaagtgga agacttaatg cctcgctgga taaatcatca 1680
ggacgtagta ctttatacat tgcagcttct cagcctggtg actcagccac ctacctctgt 1740
gctgtgaggc ccctgtacgg aggaagctac atacctacat ttggaagagg aaccagcctt 1800
attgttcatc cgtatatcca gaaccctgac cctgccgtgt accagctgag agactctaaa 1860
tccagtgaca agtctgtctg cctattcacc gattttgatt ctcaaacaaa tgtgtcacaa 1920
agtaaggatt ctgatgtgta tatcacagac aaaactgtgc tagacatgag gtctatggac 1980
ttcaagagca acagtgctgt ggcctggagc aacaaatctg actttgcatg tgcaaacgcc 2040
ttcaacaaca gcattattcc agaagacacc ttcttcccca gcccagaaag ttccggctcc 2100
ccaaaatgtg atatctacat ctgggcgccc ttggccggga cttgtggggt ccttctcctg 2160
tcactggtta tcacccttta ctgcaaacgg ggcagaaaga aactcctgta tatattcaaa 2220
caaccattta tgagaccagt acaaactact caagaggaag atggctgtag ctgccgattt 2280
ccagaagaag aagaaggagg atgtgaactg agagtgaagt tcagcaggag cgcagacgcc 2340
cccgcgtacc agcagggcca gaaccagctc tataacgagc tcaatctagg acgaagagag 2400
gagtacgatg ttttggacaa gagacgtggc cgggaccctg agatgggggg aaagccgaga 2460
aggaagaacc ctcaggaagg cctgtacaat gaactgcaga aagataagat ggcggaggcc 2520
tacagtgaga ttgggatgaa aggcgagcgc cggaggggca aggggcacga tggcctttac 2580
cagggtctca gtacagccac caaggacacc tacgacgccc ttcacatgca ggccctgccc 2640
cctcgctaa 2649
<210> 61
<211> 2898
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 61
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagacggct ctcctaagtg cgacatttat atttgggccc ctctcgctgg cacatgcggc 840
gtgttgttgc tcagcctcgt gattacactt tattgtagga gtaagaggag caggctcctg 900
cacagtgact acatgaacat gactccccgc cgccccgggc ccacccgcaa gcattaccag 960
ccctatgccc caccacgcga cttcgcagcc tatcgctccc gctcaaaacg ctctcgcttg 1020
ttacattccg attatatgaa tatgacacct aggagacctg gcccgactag gaaacactat 1080
caaccttacg cacctcccag agattttgct gcttacagga gtcgggtcaa attttcacgc 1140
tccgctgatg ctcctgccta tcaacaaggg caaaatcaat tgtacaatga attgaacttg 1200
ggtagaaggg aagaatatga cgtgctcgat aaacggaggg ggagagatcc agaaatgggc 1260
ggtaaaccac ggcgcaaaaa tccacaagag ggattgtata acgagctcca aaaggacaaa 1320
atggcagaag cttattcaga aataggaatg aagggggaaa ggagacgagg taaaggtcat 1380
gacggattgt atcaaggatt gtcaaccgct actaaagata catatgatgc tttgcatatg 1440
caagctttgc ctcccagagc caagcggtct gggtctgggg ccaccaactt cagcctgctg 1500
aagcaggccg gcgacgtgga ggagaacccc ggccccatgg agaccctctt gggcctgctt 1560
atcctttggc tgcagctgca atgggtgagc agcaaacagg aggtgacgca gattcctgca 1620
gctctgagtg tcccagaagg agaaaacttg gttctcaact gcagtttcac tgatagcgct 1680
atttacaacc tccagtggtt taggcaggac cctgggaaag gtctcacatc tctgttgctt 1740
attcagtcaa gtcagagaga gcaaacaagt ggaagactta atgcctcgct ggataaatca 1800
tcaggacgta gtactttata cattgcagct tctcagcctg gtgactcagc cacctacctc 1860
tgtgctgtga ggcccctgta cggaggaagc tacataccta catttggaag aggaaccagc 1920
cttattgttc atccgtatat ccagaaccct gaccctgccg tgtaccagct gagagactct 1980
aaatccagtg acaagtctgt ctgcctattc accgattttg attctcaaac aaatgtgtca 2040
caaagtaagg attctgatgt gtatatcaca gacaaaactg tgctagacat gaggtctatg 2100
gacttcaaga gcaacagtgc tgtggcctgg agcaacaaat ctgactttgc atgtgcaaac 2160
gccttcaaca acagcattat tccagaagac accttcttcc ccagcccaga aagttccggc 2220
tccccaaaat gtgatatcta catctgggcg cccttggccg ggacttgtgg ggtccttctc 2280
ctgtcactgg ttatcaccct ttactgcaag agagggcgta aaaagctgct ctacatcttt 2340
aagcagcctt tcatgcgtcc tgttcagaca acacaggaag aggacggatg ctcttgcagg 2400
ttccctgagg aggaggaggg tggttgcgag ctcaaacggg gcagaaagaa actcctgtat 2460
atattcaaac aaccatttat gagaccagta caaactactc aagaggaaga tggctgtagc 2520
tgccgatttc cagaagaaga agaaggagga tgtgaactga gagtgaagtt cagcaggagc 2580
gcagacgccc ccgcgtacca gcagggccag aaccagctct ataacgagct caatctagga 2640
cgaagagagg agtacgatgt tttggacaag agacgtggcc gggaccctga gatgggggga 2700
aagccgagaa ggaagaaccc tcaggaaggc ctgtacaatg aactgcagaa agataagatg 2760
gcggaggcct acagtgagat tgggatgaaa ggcgagcgcc ggaggggcaa ggggcacgat 2820
ggcctttacc agggtctcag tacagccacc aaggacacct acgacgccct tcacatgcag 2880
gccctgcccc ctcgctaa 2898
<210> 62
<211> 3180
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 62
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagacggct ctcctaaggc aaaaccgacg accacccctg cccccaggcc tcctactccc 840
gccccgacga ttgccagcca accgttaagt ttaagaccgg aagcatgtag accggcagct 900
ggtggggctg ttcatacacg tggcttagat tttgcgccta ggaagatcga ggtaatgtac 960
ccaccgccct atcttgataa cgaaaaatct aacggtacaa taattcacgt caagggcaag 1020
catttgtgcc cttccccgtt gttcccgggc ccaagcaaac cgttctgggt tctcgtcgtc 1080
gtgggaggtg tgttagcatg ttactctctc ttggttactg tcgctttcat aatcttttgg 1140
gtccgctcaa aacgctctcg cttgttacat tccgattata tgaatatgac acctaggaga 1200
cctggcccga ctaggaaaca ctatcaacct tacgcacctc ccagagattt tgctgcttac 1260
aggagtcggg tcaaattttc acgctccgct gatgctcctg cctatcaaca agggcaaaat 1320
caattgtaca atgaattgaa cttgggtaga agggaagaat atgacgtgct cgataaacgg 1380
agggggagag atccagaaat gggcggtaaa ccacggcgca aaaatccaca agagggattg 1440
tataacgagc tccaaaagga caaaatggca gaagcttatt cagaaatagg aatgaagggg 1500
gaaaggagac gaggtaaagg tcatgacgga ttgtatcaag gattgtcaac cgctactaaa 1560
gatacatatg atgctttgca tatgcaagct ttgcctccca gagccaagcg gtctgggtct 1620
ggggccacca acttcagcct gctgaagcag gccggcgacg tggaggagaa ccccggcccc 1680
atggagaccc tcttgggcct gcttatcctt tggctgcagc tgcaatgggt gagcagcaaa 1740
caggaggtga cgcagattcc tgcagctctg agtgtcccag aaggagaaaa cttggttctc 1800
aactgcagtt tcactgatag cgctatttac aacctccagt ggtttaggca ggaccctggg 1860
aaaggtctca catctctgtt gcttattcag tcaagtcaga gagagcaaac aagtggaaga 1920
cttaatgcct cgctggataa atcatcagga cgtagtactt tatacattgc agcttctcag 1980
cctggtgact cagccaccta cctctgtgct gtgaggcccc tgtacggagg aagctacata 2040
cctacatttg gaagaggaac cagccttatt gttcatccgt atatccagaa ccctgaccct 2100
gccgtgtacc agctgagaga ctctaaatcc agtgacaagt ctgtctgcct attcaccgat 2160
tttgattctc aaacaaatgt gtcacaaagt aaggattctg atgtgtatat cacagacaaa 2220
actgtgctag acatgaggtc tatggacttc aagagcaaca gtgctgtggc ctggagcaac 2280
aaatctgact ttgcatgtgc aaacgccttc aacaacagca ttattccaga agacaccttc 2340
ttccccagcc cagaaagttc cggctcccca aaagctaagc ccaccacgac gccagcgccg 2400
cgaccaccaa caccggcgcc caccatcgcg tcgcagcccc tgtccctgcg cccagaggcg 2460
tgccggccag cggcgggggg cgcagtgcac acgagggggc tggacttcgc ccctaggaaa 2520
attgaagtta tgtatcctcc tccttaccta gacaatgaga agagcaatgg aaccattatc 2580
catgtgaaag ggaaacacct ttgtccaagt cccctatttc ccggaccttc taagcccttt 2640
tgggtgctgg tggtggttgg tggagtcctg gcttgctata gcttgctagt aacagtggcc 2700
tttattattt tctgggtgag gagtaagagg agcaggctcc tgcacagtga ctacatgaac 2760
atgactcccc gccgccccgg gcccacccgc aagcattacc agccctatgc cccaccacgc 2820
gacttcgcag cctatcgctc cagagtgaag ttcagcagga gcgcagacgc ccccgcgtac 2880
cagcagggcc agaaccagct ctataacgag ctcaatctag gacgaagaga ggagtacgat 2940
gttttggaca agagacgtgg ccgggaccct gagatggggg gaaagccgag aaggaagaac 3000
cctcaggaag gcctgtacaa tgaactgcag aaagataaga tggcggaggc ctacagtgag 3060
attgggatga aaggcgagcg ccggaggggc aaggggcacg atggccttta ccagggtctc 3120
agtacagcca ccaaggacac ctacgacgcc cttcacatgc aggccctgcc ccctcgctaa 3180
<210> 63
<211> 2580
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 63
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagacggct ctcctaaggc aaaaccgacg accacccctg cccccaggcc tcctactccc 840
gccccgacga ttgccagcca accgttaagt ttaagaccgg aagcatgtag accggcagct 900
ggtggggctg ttcatacacg tggcttagat tttgcgccta ggaagatcga ggtaatgtac 960
ccaccgccct atcttgataa cgaaaaatct aacggtacaa taattcacgt caagggcaag 1020
catttgtgcc cttccccgtt gttcccgggc ccaagcaaac cgttctgggt tctcgtcgtc 1080
gtgggaggtg tgttagcatg ttactctctc ttggttactg tcgctttcat aatcttttgg 1140
gtccgctcaa aacgctctcg cttgttacat tccgattata tgaatatgac acctaggaga 1200
cctggcccga ctaggaaaca ctatcaacct tacgcacctc ccagagattt tgctgcttac 1260
aggagtcggg tcaaattttc acgctccgct gatgctcctg cctatcaaca agggcaaaat 1320
caattgtaca atgaattgaa cttgggtaga agggaagaat atgacgtgct cgataaacgg 1380
agggggagag atccagaaat gggcggtaaa ccacggcgca aaaatccaca agagggattg 1440
tataacgagc tccaaaagga caaaatggca gaagcttatt cagaaatagg aatgaagggg 1500
gaaaggagac gaggtaaagg tcatgacgga ttgtatcaag gattgtcaac cgctactaaa 1560
gatacatatg atgctttgca tatgcaagct ttgcctccca gagccaagcg gtctgggtct 1620
ggggccacca acttcagcct gctgaagcag gccggcgacg tggaggagaa ccccggcccc 1680
atggagaccc tcttgggcct gcttatcctt tggctgcagc tgcaatgggt gagcagcaaa 1740
caggaggtga cgcagattcc tgcagctctg agtgtcccag aaggagaaaa cttggttctc 1800
aactgcagtt tcactgatag cgctatttac aacctccagt ggtttaggca ggaccctggg 1860
aaaggtctca catctctgtt gcttattcag tcaagtcaga gagagcaaac aagtggaaga 1920
cttaatgcct cgctggataa atcatcagga cgtagtactt tatacattgc agcttctcag 1980
cctggtgact cagccaccta cctctgtgct gtgaggcccc tgtacggagg aagctacata 2040
cctacatttg gaagaggaac cagccttatt gttcatccgt atatccagaa ccctgaccct 2100
gccgtgtacc agctgagaga ctctaaatcc agtgacaagt ctgtctgcct attcaccgat 2160
tttgattctc aaacaaatgt gtcacaaagt aaggattctg atgtgtatat cacagacaaa 2220
actgtgctag acatgaggtc tatggacttc aagagcaaca gtgctgtggc ctggagcaac 2280
aaatctgact ttgcatgtgc aaacgccttc aacaacagca ttattccaga agacaccttc 2340
ttccccagcc cagaaagttc cggctcccca aaatgtgata tctacatctg ggcgcccttg 2400
gccgggactt gtggggtcct tctcctgtca ctggttatca ccctttactg caaacggggc 2460
agaaagaaac tcctgtatat attcaaacaa ccatttatga gaccagtaca aactactcaa 2520
gaggaagatg gctgtagctg ccgatttcca gaagaagaag aaggaggatg tgaactgtaa 2580
<210> 64
<211> 612
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 64
Met Ser Ile Gly Leu Leu Cys Cys Ala Ala Leu Ser Leu Leu Trp Ala
1 5 10 15
Gly Pro Val Asn Ala Gly Val Thr Gln Thr Pro Lys Phe Gln Val Leu
20 25 30
Lys Thr Gly Gln Ser Met Thr Leu Gln Cys Ala Gln Asp Met Asn His
35 40 45
Glu Tyr Met Ser Trp Tyr Arg Gln Asp Pro Gly Met Gly Leu Arg Leu
50 55 60
Ile His Tyr Ser Val Gly Ala Gly Ile Thr Asp Gln Gly Glu Val Pro
65 70 75 80
Asn Gly Tyr Asn Val Ser Arg Ser Thr Thr Glu Asp Phe Pro Leu Arg
85 90 95
Leu Leu Ser Ala Ala Pro Ser Gln Thr Ser Val Tyr Phe Cys Ala Ser
100 105 110
Ser Tyr Val Gly Asn Thr Gly Glu Leu Phe Phe Gly Glu Gly Ser Arg
115 120 125
Leu Thr Val Leu Glu Asp Leu Lys Asn Val Phe Pro Pro Glu Val Ala
130 135 140
Val Phe Glu Pro Ser Glu Ala Glu Ile Ser His Thr Gln Lys Ala Thr
145 150 155 160
Leu Val Cys Leu Ala Thr Gly Phe Tyr Pro Asp His Val Glu Leu Ser
165 170 175
Trp Trp Val Asn Gly Lys Glu Val His Ser Gly Val Ser Thr Asp Pro
180 185 190
Gln Pro Leu Lys Glu Gln Pro Ala Leu Asn Asp Ser Arg Tyr Ala Leu
195 200 205
Ser Ser Arg Leu Arg Val Ser Ala Thr Phe Trp Gln Asp Pro Arg Asn
210 215 220
His Phe Arg Cys Gln Val Gln Phe Tyr Gly Leu Ser Glu Asn Asp Glu
225 230 235 240
Trp Thr Gln Asp Arg Ala Lys Pro Val Thr Gln Ile Val Ser Ala Glu
245 250 255
Ala Trp Gly Arg Ala Asp Cys Gly Phe Thr Ser Glu Ser Tyr Gln Gln
260 265 270
Gly Val Leu Ser Ala Thr Ile Leu Tyr Glu Ile Leu Leu Gly Lys Ala
275 280 285
Thr Leu Tyr Ala Val Leu Val Ser Ala Leu Val Leu Met Ala Met Val
290 295 300
Lys Arg Lys Asp Ser Arg Gly Ala Lys Arg Ser Gly Ser Gly Ala Thr
305 310 315 320
Asn Phe Ser Leu Leu Lys Gln Ala Gly Asp Val Glu Glu Asn Pro Gly
325 330 335
Pro Met Glu Thr Leu Leu Gly Leu Leu Ile Leu Trp Leu Gln Leu Gln
340 345 350
Trp Val Ser Ser Lys Gln Glu Val Thr Gln Ile Pro Ala Ala Leu Ser
355 360 365
Val Pro Glu Gly Glu Asn Leu Val Leu Asn Cys Ser Phe Thr Asp Ser
370 375 380
Ala Ile Tyr Asn Leu Gln Trp Phe Arg Gln Asp Pro Gly Lys Gly Leu
385 390 395 400
Thr Ser Leu Leu Leu Ile Gln Ser Ser Gln Arg Glu Gln Thr Ser Gly
405 410 415
Arg Leu Asn Ala Ser Leu Asp Lys Ser Ser Gly Arg Ser Thr Leu Tyr
420 425 430
Ile Ala Ala Ser Gln Pro Gly Asp Ser Ala Thr Tyr Leu Cys Ala Val
435 440 445
Arg Pro Leu Tyr Gly Gly Ser Tyr Ile Pro Thr Phe Gly Arg Gly Thr
450 455 460
Ser Leu Ile Val His Pro Tyr Ile Gln Asn Pro Asp Pro Ala Val Tyr
465 470 475 480
Gln Leu Arg Asp Ser Lys Ser Ser Asp Lys Ser Val Cys Leu Phe Thr
485 490 495
Asp Phe Asp Ser Gln Thr Asn Val Ser Gln Ser Lys Asp Ser Asp Val
500 505 510
Tyr Ile Thr Asp Lys Thr Val Leu Asp Met Arg Ser Met Asp Phe Lys
515 520 525
Ser Asn Ser Ala Val Ala Trp Ser Asn Lys Ser Asp Phe Ala Cys Ala
530 535 540
Asn Ala Phe Asn Asn Ser Ile Ile Pro Glu Asp Thr Phe Phe Pro Ser
545 550 555 560
Pro Glu Ser Ser Cys Asp Val Lys Leu Val Glu Lys Ser Phe Glu Thr
565 570 575
Asp Thr Asn Leu Asn Phe Gln Asn Leu Ser Val Ile Gly Phe Arg Ile
580 585 590
Leu Leu Leu Lys Val Ala Gly Phe Asn Leu Leu Met Thr Leu Arg Leu
595 600 605
Trp Ser Ser Arg
610
<210> 65
<211> 1839
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 65
atgagcatcg gcctcctgtg ctgtgcagcc ttgtctctcc tgtgggcagg tccagtgaat 60
gctggtgtca ctcagacccc aaaattccag gtcctgaaga caggacagag catgacactg 120
cagtgtgccc aggatatgaa ccatgaatac atgtcctggt atcgacaaga cccaggcatg 180
gggctgaggc tgattcatta ctcagttggt gctggtatca ctgaccaagg agaagtcccc 240
aatggctaca atgtctccag atcaaccaca gaggatttcc cgctcaggct gctgtcggct 300
gctccctccc agacatctgt gtacttctgt gccagcagtt acgtcgggaa caccggggag 360
ctgttttttg gagaaggctc taggctgacc gtactggagg acctgaaaaa cgtgttccca 420
cccgaggtcg ctgtgtttga gccatcagaa gcagagatct cccacaccca aaaggccaca 480
ctggtgtgcc tggccacagg cttctacccc gaccacgtgg agctgagctg gtgggtgaat 540
gggaaggagg tgcacagtgg ggtcagcaca gacccgcagc ccctcaagga gcagcccgcc 600
ctcaatgact ccagatacgc tctgagcagc cgcctgaggg tctcggccac cttctggcag 660
gacccccgca accacttccg ctgtcaagtc cagttctacg ggctctcgga gaatgacgag 720
tggacccagg atagggccaa acccgtcacc cagatcgtca gcgccgaggc ctggggtaga 780
gcagactgtg gcttcacctc cgagtcttac cagcaagggg tcctgtctgc caccatcctc 840
tatgagatct tgctagggaa ggccaccttg tatgccgtgc tggtcagtgc cctcgtgctg 900
atggctatgg tcaagagaaa ggattccaga ggcgccaagc ggtctgggtc tggggccacc 960
aacttcagcc tgctgaagca ggccggcgac gtggaggaga accccggccc catggagacc 1020
ctcttgggcc tgcttatcct ttggctgcag ctgcaatggg tgagcagcaa acaggaggtg 1080
acgcagattc ctgcagctct gagtgtccca gaaggagaaa acttggttct caactgcagt 1140
ttcactgata gcgctattta caacctccag tggtttaggc aggaccctgg gaaaggtctc 1200
acatctctgt tgcttattca gtcaagtcag agagagcaaa caagtggaag acttaatgcc 1260
tcgctggata aatcatcagg acgtagtact ttatacattg cagcttctca gcctggtgac 1320
tcagccacct acctctgtgc tgtgaggccc ctgtacggag gaagctacat acctacattt 1380
ggaagaggaa ccagccttat tgttcatccg tatatccaga accctgaccc tgccgtgtac 1440
cagctgagag actctaaatc cagtgacaag tctgtctgcc tattcaccga ttttgattct 1500
caaacaaatg tgtcacaaag taaggattct gatgtgtata tcacagacaa aactgtgcta 1560
gacatgaggt ctatggactt caagagcaac agtgctgtgg cctggagcaa caaatctgac 1620
tttgcatgtg caaacgcctt caacaacagc attattccag aagacacctt cttccccagc 1680
ccagaaagtt cctgtgatgt caagctggtc gagaaaagct ttgaaacaga tacgaaccta 1740
aactttcaaa acctgtcagt gattgggttc cgaatcctcc tcctgaaagt ggccgggttt 1800
aatctgctca tgacgctgcg gctgtggtcc agccggtaa 1839
<210> 66
<211> 48
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 66
Cys Asp Val Lys Leu Val Glu Lys Ser Phe Glu Thr Asp Thr Asn Leu
1 5 10 15
Asn Phe Gln Asn Leu Ser Val Ile Gly Phe Arg Ile Leu Leu Leu Lys
20 25 30
Val Ala Gly Phe Asn Leu Leu Met Thr Leu Arg Leu Trp Ser Ser Arg
35 40 45
<210> 67
<211> 7
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 67
Ala Lys Arg Ser Gly Ser Gly
1 5
<210> 68
<211> 49
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 68
Cys Gly Phe Thr Ser Glu Ser Tyr Gln Gln Gly Val Leu Ser Ala Thr
1 5 10 15
Ile Leu Tyr Glu Ile Leu Leu Gly Lys Ala Thr Leu Tyr Ala Val Leu
20 25 30
Val Ser Ala Leu Val Leu Met Ala Met Val Lys Arg Lys Asp Ser Arg
35 40 45
Gly
<210> 69
<211> 4
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 69
Gly Ser Pro Lys
1
<210> 70
<211> 2
<212> PRT
<213> Artificial sequence
<220>
<223> synthetic
<400> 70
Pro Arg
1
<210> 71
<211> 6
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 71
ctcgag 6
<210> 72
<211> 14
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 72
cagccagcgg ccgc 14
<210> 73
<211> 8
<212> DNA
<213> Artificial sequence
<220>
<223> synthetic
<400> 73
gcggccgc 8
- 上一篇:一种医用注射器针头装配设备
- 下一篇:具有增强的抗体导向免疫应答的人自然杀伤细胞亚群
